
| Name: LOC100291514 | Sequence: fasta or formatted (250aa) | NCBI GI: 239749793 | |
|
Description: PREDICTED: hypothetical protein XP_002347163
| Not currently referenced in the text | ||
|
Composition:
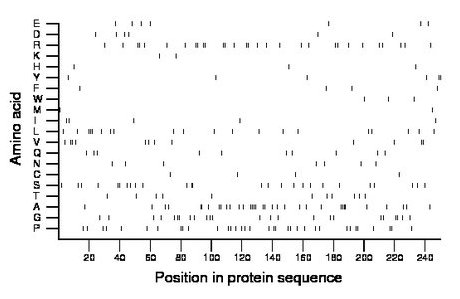
Amino acid Percentage Count Longest homopolymer A alanine 13.6 34 4 C cysteine 1.6 4 1 D aspartate 2.4 6 1 E glutamate 2.8 7 1 F phenylalanine 1.6 4 1 G glycine 9.6 24 2 H histidine 1.2 3 1 I isoleucine 2.0 5 1 K lysine 0.8 2 1 L leucine 8.0 20 3 M methionine 0.8 2 1 N asparagine 2.8 7 1 P proline 13.6 34 2 Q glutamine 4.0 10 1 R arginine 12.0 30 2 S serine 10.4 26 2 T threonine 4.4 11 1 V valine 4.8 12 2 W tryptophan 1.2 3 1 Y tyrosine 2.4 6 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347163 LOC100289311 1.000 PREDICTED: hypothetical protein XP_002343026 LOC100294010 0.949 PREDICTED: hypothetical protein SCARF2 0.058 scavenger receptor class F, member 2 isoform 1 [Homo... SCARF2 0.058 scavenger receptor class F, member 2 isoform 2 [Homo... SYN1 0.049 synapsin I isoform Ia SYN1 0.049 synapsin I isoform Ib TSC22D1 0.041 TSC22 domain family, member 1 isoform 1 FLJ22184 0.039 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.039 PREDICTED: hypothetical protein LOC80164 LOC100289661 0.039 PREDICTED: hypothetical protein LOC100289661 0.039 PREDICTED: hypothetical protein XP_002343723 LOC728650 0.039 PREDICTED: hypothetical protein TAF4 0.039 TBP-associated factor 4 MAVS 0.037 virus-induced signaling adapter UNCX 0.037 UNC homeobox LOC100292172 0.037 PREDICTED: hypothetical protein UTF1 0.037 undifferentiated embryonic cell transcription factor... FLJ31945 0.035 PREDICTED: hypothetical protein LOC100293818 0.035 PREDICTED: hypothetical protein LOC100131102 0.035 PREDICTED: hypothetical protein LOC100131102 0.035 PREDICTED: hypothetical protein SHANK3 0.035 SH3 and multiple ankyrin repeat domains 3 FAM48B1 0.035 hypothetical protein LOC100130302 AGAP2 0.035 centaurin, gamma 1 isoform PIKE-L LOC729580 0.035 PREDICTED: hypothetical protein LOC729580 0.035 PREDICTED: hypothetical protein LOC729580 0.035 PREDICTED: hypothetical protein APC2 0.035 adenomatosis polyposis coli 2 MEX3A 0.035 MEX3A proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
