
| Name: HGC6.3 | Sequence: fasta or formatted (171aa) | NCBI GI: 193804858 | |
|
Description: hypothetical protein LOC100128124
| Not currently referenced in the text | ||
|
Composition:
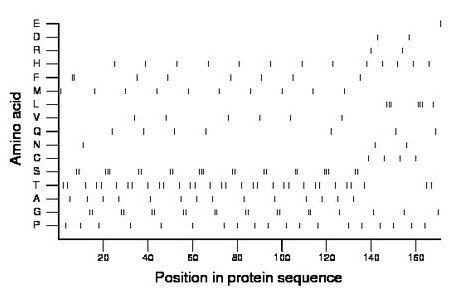
Amino acid Percentage Count Longest homopolymer A alanine 8.2 14 1 C cysteine 2.3 4 1 D aspartate 1.2 2 1 E glutamate 0.6 1 1 F phenylalanine 4.7 8 2 G glycine 11.7 20 2 H histidine 7.6 13 1 I isoleucine 0.0 0 0 K lysine 0.0 0 0 L leucine 4.1 7 3 M methionine 5.8 10 1 N asparagine 1.8 3 1 P proline 11.1 19 1 Q glutamine 4.1 7 1 R arginine 1.2 2 1 S serine 13.5 23 3 T threonine 18.7 32 1 V valine 3.5 6 1 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC100128124 POLR2A 0.095 DNA-directed RNA polymerase II A HDAC6 0.083 histone deacetylase 6 C11orf24 0.074 hypothetical protein LOC53838 MUC5AC 0.068 mucin 5AC MUC5B 0.068 mucin 5, subtype B, tracheobronchial LOC100294420 0.062 PREDICTED: similar to mucin 2 SRRM2 0.056 splicing coactivator subunit SRm300 MUC12 0.056 PREDICTED: mucin 12 LOC100130716 0.053 PREDICTED: similar to mucin 11 LOC100130716 0.053 PREDICTED: similar to Mucin-12 FAM65A 0.053 hypothetical protein LOC79567 MUC6 0.053 mucin 6, gastric TGOLN2 0.053 trans-golgi network protein 2 LOC100133915 0.050 PREDICTED: hypothetical protein MUC4 0.050 mucin 4 isoform a RBM16 0.050 RNA-binding motif protein 16 MUC7 0.050 mucin 7, secreted precursor LOC100132635 0.050 PREDICTED: similar to mucin 5, partial MUC7 0.050 mucin 7, secreted precursor MUC7 0.050 mucin 7, secreted precursor POM121C 0.050 POM121 membrane glycoprotein (rat)-like MUC3A 0.047 PREDICTED: mucin 3A, intestinal LOC732156 0.047 PREDICTED: similar to Mucin-3A LOC100133761 0.047 PREDICTED: similar to Mucin-6, partial ZAN 0.044 zonadhesin isoform 3 ZAN 0.044 zonadhesin isoform 6 LOC100288540 0.044 PREDICTED: hypothetical protein XP_002342775 POM121 0.044 nuclear pore membrane protein 121 LOC100293485 0.044 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
