
| Name: LOC100288662 | Sequence: fasta or formatted (85aa) | NCBI GI: 239741567 | |
|
Description: PREDICTED: hypothetical protein XP_002342238
| Not currently referenced in the text | ||
|
Composition:
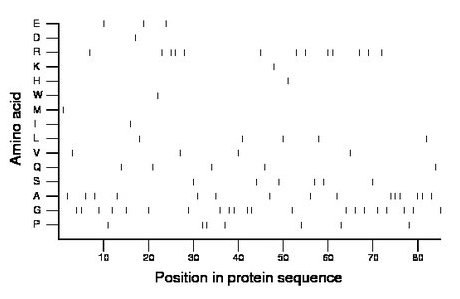
Amino acid Percentage Count Longest homopolymer A alanine 17.6 15 3 C cysteine 0.0 0 0 D aspartate 1.2 1 1 E glutamate 3.5 3 1 F phenylalanine 0.0 0 0 G glycine 24.7 21 2 H histidine 1.2 1 1 I isoleucine 1.2 1 1 K lysine 1.2 1 1 L leucine 5.9 5 1 M methionine 1.2 1 1 N asparagine 0.0 0 0 P proline 8.2 7 2 Q glutamine 5.9 5 1 R arginine 15.3 13 2 S serine 7.1 6 1 T threonine 0.0 0 0 V valine 4.7 4 1 W tryptophan 1.2 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342238 LOC100292412 1.000 PREDICTED: hypothetical protein ALPK3 0.095 alpha-kinase 3 LOC100293136 0.088 PREDICTED: hypothetical protein LOC100290831 0.088 PREDICTED: hypothetical protein XP_002347799 LOC100287562 0.088 PREDICTED: hypothetical protein XP_002343591 GSK3A 0.081 glycogen synthase kinase 3 alpha COL1A1 0.081 alpha 1 type I collagen preproprotein LOC100291076 0.074 PREDICTED: hypothetical protein XP_002347846 DACT3 0.074 thymus expressed gene 3-like LOC100130819 0.068 PREDICTED: hypothetical protein ANKRD56 0.068 ankyrin repeat domain 56 LOC100130819 0.061 PREDICTED: hypothetical protein LOC651986 0.061 PREDICTED: similar to forkhead box L2 LOC100130819 0.061 PREDICTED: hypothetical protein LOC100288524 0.061 PREDICTED: hypothetical protein XP_002342748 LOC651986 0.061 PREDICTED: similar to forkhead box L2 SYNCRIP 0.061 synaptotagmin binding, cytoplasmic RNA interacting ... SYNCRIP 0.061 synaptotagmin binding, cytoplasmic RNA interacting ... FGF2 0.054 fibroblast growth factor 2 LOC728650 0.054 PREDICTED: hypothetical protein LOC728650 0.054 PREDICTED: hypothetical protein LOC100287290 0.054 PREDICTED: hypothetical protein LOC100130897 0.054 PREDICTED: hypothetical protein LOC100287290 0.054 PREDICTED: hypothetical protein XP_002342446 LOC100130897 0.054 PREDICTED: hypothetical protein COL7A1 0.054 alpha 1 type VII collagen precursor MAGI2 0.054 membrane associated guanylate kinase, WW and PDZ dom... SYNCRIP 0.047 synaptotagmin binding, cytoplasmic RNA interacting ... LOC100132234 0.047 PREDICTED: hypothetical protein LOC100132234Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
