
| Name: LOC100287437 | Sequence: fasta or formatted (144aa) | NCBI GI: 239741134 | |
|
Description: PREDICTED: hypothetical protein XP_002342117
| Not currently referenced in the text | ||
|
Composition:
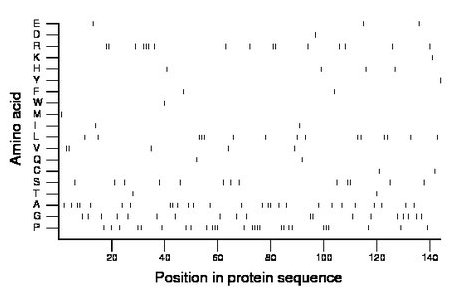
Amino acid Percentage Count Longest homopolymer A alanine 18.8 27 2 C cysteine 1.4 2 1 D aspartate 0.7 1 1 E glutamate 2.1 3 1 F phenylalanine 1.4 2 1 G glycine 13.2 19 2 H histidine 2.8 4 1 I isoleucine 1.4 2 1 K lysine 0.7 1 1 L leucine 10.4 15 3 M methionine 0.7 1 1 N asparagine 0.0 0 0 P proline 18.8 27 4 Q glutamine 1.4 2 1 R arginine 11.1 16 3 S serine 9.0 13 2 T threonine 1.4 2 1 V valine 3.5 5 2 W tryptophan 0.7 1 1 Y tyrosine 0.7 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342117 LOC100290551 1.000 PREDICTED: hypothetical protein XP_002346417 SAMD1 0.110 sterile alpha motif domain containing 1 CASKIN1 0.103 CASK interacting protein 1 TAF4 0.095 TBP-associated factor 4 LOC390595 0.084 PREDICTED: similar to ubiquitin-associated protein ... LOC390595 0.084 PREDICTED: similar to ubiquitin-associated protein ... LOC390595 0.084 PREDICTED: hypothetical protein DCAF10 0.081 WD repeat domain 32 WIPF2 0.081 WIRE protein C6orf132 0.077 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.077 PREDICTED: chromosome 6 open reading frame 132 [Hom... C6orf132 0.077 PREDICTED: hypothetical protein LOC647024 RANBP9 0.077 RAN binding protein 9 WASF2 0.073 WAS protein family, member 2 FMN2 0.073 formin 2 SF3A2 0.073 splicing factor 3a, subunit 2 WASL 0.070 Wiskott-Aldrich syndrome gene-like protein C10orf95 0.070 hypothetical protein LOC79946 ADAM15 0.070 a disintegrin and metalloproteinase domain 15 isofor... WIPF3 0.070 WAS/WASL interacting protein family, member 3 [Homo... LOC100131102 0.070 PREDICTED: hypothetical protein LOC100131102 0.070 PREDICTED: hypothetical protein MLL 0.066 myeloid/lymphoid or mixed-lineage leukemia protein [... ADAM15 0.066 a disintegrin and metalloproteinase domain 15 isofor... IRX5 0.066 iroquois homeobox protein 5 WBP11 0.066 WW domain binding protein 11 BBC3 0.062 BCL2 binding component 3 isoform 1 GLTSCR1 0.062 glioma tumor suppressor candidate region gene 1 [Ho... DIAPH1 0.062 diaphanous 1 isoform 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
