
| Name: C20orf112 | Sequence: fasta or formatted (436aa) | NCBI GI: 193788697 | |
|
Description: hypothetical protein LOC140688
|
Referenced in:
| ||
|
Composition:
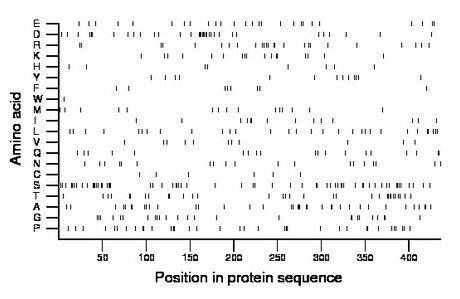
Amino acid Percentage Count Longest homopolymer A alanine 8.5 37 3 C cysteine 1.4 6 1 D aspartate 7.8 34 6 E glutamate 6.0 26 1 F phenylalanine 1.8 8 1 G glycine 5.7 25 2 H histidine 1.8 8 1 I isoleucine 3.2 14 2 K lysine 3.9 17 2 L leucine 6.9 30 2 M methionine 3.4 15 1 N asparagine 4.4 19 1 P proline 8.0 35 2 Q glutamine 4.4 19 2 R arginine 5.7 25 2 S serine 14.0 61 3 T threonine 7.3 32 2 V valine 2.5 11 1 W tryptophan 0.2 1 1 Y tyrosine 3.0 13 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC140688 NOL4 0.434 nucleolar protein 4 DSPP 0.021 dentin sialophosphoprotein preproprotein CHD1 0.019 chromodomain helicase DNA binding protein 1 NAP1L3 0.016 nucleosome assembly protein 1-like 3 LOC100128719 0.015 PREDICTED: hypothetical protein, partial LOC100128719 0.015 PREDICTED: hypothetical protein, partial TOX3 0.014 TOX high mobility group box family member 3 isoform... TOX3 0.014 TOX high mobility group box family member 3 isoform... LOC100134147 0.014 PREDICTED: hypothetical protein LOC100294420 0.014 PREDICTED: similar to mucin 2 TNRC18 0.013 trinucleotide repeat containing 18 HNRNPUL1 0.013 heterogeneous nuclear ribonucleoprotein U-like 1 iso... HNRNPUL1 0.013 heterogeneous nuclear ribonucleoprotein U-like 1 iso... IRS2 0.013 insulin receptor substrate 2 NHSL1 0.012 NHS-like 1 MAGEE1 0.012 melanoma antigen family E, 1 SRRM2 0.012 splicing coactivator subunit SRm300 CASQ2 0.012 cardiac calsequestrin 2 precursor LOC100133790 0.012 PREDICTED: intestinal mucin-like, partial COL27A1 0.012 collagen, type XXVII, alpha 1 GGA1 0.012 golgi associated, gamma adaptin ear containing, ARF ... DZIP1L 0.012 DAZ interacting protein 1-like RNF34 0.011 ring finger protein 34 isoform 1 RNF34 0.011 ring finger protein 34 isoform 2 MTSS1 0.011 metastasis suppressor 1 GPRIN3 0.011 G protein-regulated inducer of neurite outgrowth 3 ... STIM1 0.011 stromal interaction molecule 1 precursor NPM1 0.011 nucleophosmin 1 isoform 2 ATAD2 0.011 ATPase family, AAA domain containing 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
