
| Name: LOC100133749 | Sequence: fasta or formatted (131aa) | NCBI GI: 169216121 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
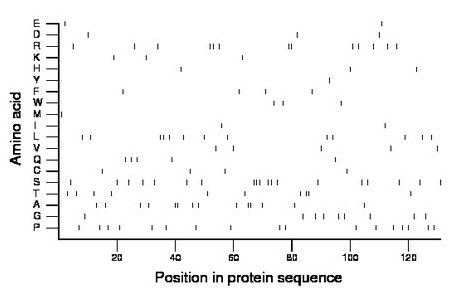
Amino acid Percentage Count Longest homopolymer A alanine 10.7 14 2 C cysteine 3.1 4 1 D aspartate 2.3 3 1 E glutamate 1.5 2 1 F phenylalanine 3.1 4 1 G glycine 6.9 9 1 H histidine 2.3 3 1 I isoleucine 1.5 2 1 K lysine 2.3 3 1 L leucine 9.9 13 2 M methionine 0.8 1 1 N asparagine 0.0 0 0 P proline 13.0 17 1 Q glutamine 3.8 5 1 R arginine 9.9 13 2 S serine 14.5 19 3 T threonine 7.6 10 2 V valine 3.8 5 1 W tryptophan 2.3 3 1 Y tyrosine 0.8 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290683 0.044 PREDICTED: hypothetical protein XP_002348231 LOC441239 0.044 PREDICTED: hypothetical protein LOC441239 LOC441239 0.040 PREDICTED: hypothetical protein LOC441239 LOC441239 0.040 PREDICTED: hypothetical protein LOC441239 LOC441239 0.032 PREDICTED: hypothetical protein LOC441239 LOC729660 0.028 PREDICTED: similar to Putative uncharacterized prot... LOC100292425 0.020 PREDICTED: hypothetical protein isoform 2 LOC100292865 0.020 PREDICTED: hypothetical protein LOC100291013 0.020 PREDICTED: hypothetical protein XP_002347574 isofor... ZDHHC5 0.020 zinc finger, DHHC domain containing 5 DSCR6 0.016 Down syndrome critical region protein 6 IRX2 0.016 iroquois homeobox 2 IRX2 0.016 iroquois homeobox 2 FAM123B 0.012 family with sequence similarity 123B LOC100130599 0.012 PREDICTED: hypothetical protein LOC100291552 0.012 PREDICTED: hypothetical protein XP_002346560 LOC100130599 0.012 PREDICTED: similar to Keratin-associated protein 10... PHF12 0.012 PHD finger protein 12 isoform 1 DALRD3 0.012 DALR anticodon binding domain containing 3 isoform 1... PHF12 0.012 PHD finger protein 12 isoform 2 ADAMTS10 0.012 ADAM metallopeptidase with thrombospondin type 1 mot... APC2 0.012 adenomatosis polyposis coli 2 AGRN 0.012 agrin IGDCC3 0.012 putative neuronal cell adhesion molecule ADAMTS7 0.008 ADAM metallopeptidase with thrombospondin type 1 mot... TSKS 0.008 testis-specific kinase substrate NOTO 0.008 notochord homeobox GPR113 0.008 G-protein coupled receptor 113 isoform 1 LOC100291627 0.008 PREDICTED: hypothetical protein XP_002345251Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
