
| Name: FAM174B | Sequence: fasta or formatted (159aa) | NCBI GI: 150170694 | |
|
Description: hypothetical protein LOC400451
| Not currently referenced in the text | ||
|
Composition:
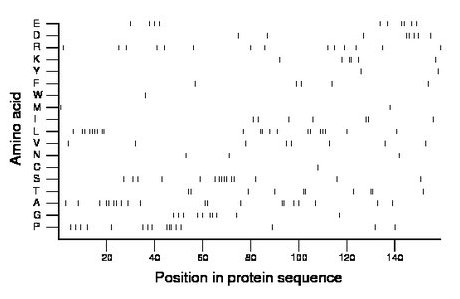
Amino acid Percentage Count Longest homopolymer A alanine 12.6 20 2 C cysteine 0.6 1 1 D aspartate 5.0 8 2 E glutamate 6.3 10 2 F phenylalanine 3.1 5 1 G glycine 6.3 10 2 H histidine 0.0 0 0 I isoleucine 4.4 7 2 K lysine 3.8 6 2 L leucine 13.2 21 4 M methionine 1.3 2 1 N asparagine 1.9 3 1 P proline 10.1 16 3 Q glutamine 0.0 0 0 R arginine 8.8 14 1 S serine 9.4 15 4 T threonine 6.3 10 2 V valine 5.0 8 1 W tryptophan 0.6 1 1 Y tyrosine 1.3 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC400451 FAM174A 0.157 family with sequence similarity 174, member A FOXK1 0.066 forkhead box K1 CNOT3 0.045 CCR4-NOT transcription complex, subunit 3 ATXN2 0.038 ataxin 2 TAF4 0.035 TBP-associated factor 4 C19orf24 0.035 hypothetical protein LOC55009 GRIN2D 0.035 N-methyl-D-aspartate receptor subunit 2D precursor ... BAT2 0.035 HLA-B associated transcript-2 UBQLN2 0.035 ubiquilin 2 KIAA1522 0.035 hypothetical protein LOC57648 SYDE1 0.035 synapse defective 1, Rho GTPase, homolog 1 LOC100132345 0.031 PREDICTED: hypothetical protein BPTF 0.031 bromodomain PHD finger transcription factor isoform ... BPTF 0.031 bromodomain PHD finger transcription factor isoform ... IRS1 0.031 insulin receptor substrate 1 LOC100127891 0.031 PREDICTED: hypothetical protein LOC100129654 0.031 PREDICTED: similar to transcription factor 23 [Homo... LOC100129654 0.031 PREDICTED: similar to transcription factor 23 [Homo... BOD1L 0.031 biorientation of chromosomes in cell division 1-like... SOX3 0.031 SRY (sex determining region Y)-box 3 LOC100294312 0.028 PREDICTED: hypothetical protein XP_002344082 LOC100294220 0.028 PREDICTED: hypothetical protein LOC100294184 0.028 PREDICTED: hypothetical protein IGFBP2 0.028 insulin-like growth factor binding protein 2, 36kDa ... HTRA1 0.028 HtrA serine peptidase 1 precursor SCAF1 0.028 SR-related CTD-associated factor 1 DMKN 0.028 dermokine isoform 4 precursor DMKN 0.028 dermokine isoform 5 precursor DMKN 0.028 dermokine isoform 3 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
