
| Name: CREBZF | Sequence: fasta or formatted (354aa) | NCBI GI: 88900495 | |
|
Description: HCF-binding transcription factor Zhangfei
|
Referenced in:
| ||
|
Composition:
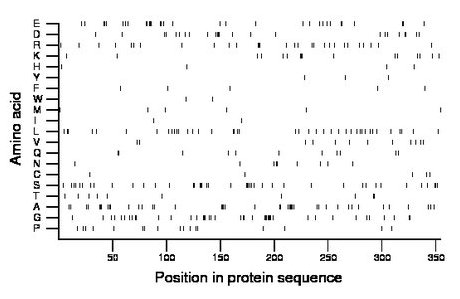
Amino acid Percentage Count Longest homopolymer A alanine 11.9 42 6 C cysteine 1.4 5 1 D aspartate 6.8 24 4 E glutamate 9.0 32 3 F phenylalanine 1.4 5 1 G glycine 11.9 42 7 H histidine 1.1 4 1 I isoleucine 0.6 2 1 K lysine 4.5 16 3 L leucine 11.6 41 2 M methionine 1.7 6 1 N asparagine 2.5 9 2 P proline 4.8 17 1 Q glutamine 3.1 11 2 R arginine 8.2 29 3 S serine 11.9 42 4 T threonine 2.8 10 2 V valine 3.4 12 1 W tryptophan 0.6 2 1 Y tyrosine 0.8 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 HCF-binding transcription factor Zhangfei ATF6B 0.037 activating transcription factor 6 beta isoform b [H... ATF6B 0.037 activating transcription factor 6 beta isoform a [Ho... CREB3L4 0.036 cAMP responsive element binding protein 3-like 4 [Ho... CCDC6 0.034 coiled-coil domain containing 6 ANKRD24 0.031 ankyrin repeat domain 24 CREB3L3 0.030 cAMP responsive element binding protein 3-like 3 [Ho... ATF6 0.027 activating transcription factor 6 SMTN 0.027 smoothelin isoform b SMTN 0.027 smoothelin isoform c SMTN 0.027 smoothelin isoform a C6orf174 0.025 hypothetical protein LOC387104 TTBK1 0.024 tau tubulin kinase 1 MN1 0.022 meningioma 1 MXRA7 0.022 transmembrane anchor protein 1 isoform 3 MXRA7 0.022 transmembrane anchor protein 1 isoform 2 MXRA7 0.022 transmembrane anchor protein 1 isoform 1 PABPN1 0.022 poly(A) binding protein, nuclear 1 TNRC18 0.021 trinucleotide repeat containing 18 CEBPG 0.021 CCAAT/enhancer binding protein gamma ARID1B 0.021 AT rich interactive domain 1B (SWI1-like) isoform 2 ... ARID1B 0.021 AT rich interactive domain 1B (SWI1-like) isoform 1 ... ARID1B 0.021 AT rich interactive domain 1B (SWI1-like) isoform 3 ... SIRT1 0.019 sirtuin 1 isoform a NPAS3 0.019 neuronal PAS domain protein 3 isoform 2 NPAS3 0.019 neuronal PAS domain protein 3 isoform 1 LOC730060 0.019 PREDICTED: hypothetical protein LOC730060 0.019 PREDICTED: hypothetical protein LOC730060 0.019 PREDICTED: hypothetical protein DACT3 0.018 thymus expressed gene 3-likeHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
