
| Name: FAM163A | Sequence: fasta or formatted (167aa) | NCBI GI: 28316806 | |
|
Description: hypothetical protein MGC16664
| Not currently referenced in the text | ||
|
Composition:
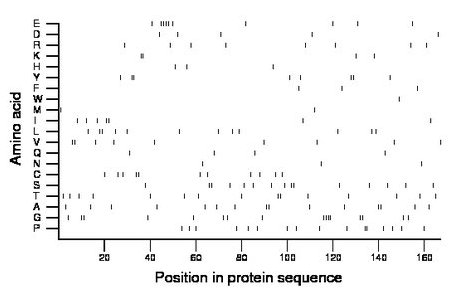
Amino acid Percentage Count Longest homopolymer A alanine 9.0 15 2 C cysteine 6.6 11 2 D aspartate 3.0 5 1 E glutamate 6.0 10 4 F phenylalanine 1.8 3 1 G glycine 9.6 16 4 H histidine 1.8 3 1 I isoleucine 4.2 7 2 K lysine 2.4 4 2 L leucine 7.2 12 2 M methionine 1.2 2 1 N asparagine 1.8 3 1 P proline 9.6 16 2 Q glutamine 2.4 4 1 R arginine 4.8 8 1 S serine 9.0 15 2 T threonine 9.0 15 2 V valine 5.4 9 2 W tryptophan 0.6 1 1 Y tyrosine 4.8 8 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein MGC16664 LOC643750 0.261 PREDICTED: hypothetical protein FAM163B 0.261 hypothetical LOC642968 RP1L1 0.021 retinitis pigmentosa 1-like 1 ATN1 0.021 atrophin-1 ATN1 0.021 atrophin-1 ANKRD11 0.021 ankyrin repeat domain 11 TSHZ1 0.018 teashirt family zinc finger 1 NBPF10 0.018 hypothetical protein LOC100132406 FRAS1 0.018 Fraser syndrome 1 NELL1 0.015 nel-like 1 isoform 2 precursor NELL1 0.015 nel-like 1 isoform 1 precursor LOC728318 0.015 PREDICTED: hypothetical protein LOC728318 0.015 PREDICTED: hypothetical protein PCDH7 0.015 protocadherin 7 isoform b precursor PCDH7 0.015 protocadherin 7 isoform c precursor PCDH7 0.015 protocadherin 7 isoform a precursor C11orf87 0.015 hypothetical protein LOC399947 DCC 0.012 deleted in colorectal carcinoma LOC100288104 0.012 PREDICTED: hypothetical protein XP_002342450 NKX2-8 0.012 NK2 homeobox 8 ZAN 0.012 zonadhesin isoform 3 KAT2B 0.009 K(lysine) acetyltransferase 2B ZFHX2 0.009 PREDICTED: zinc finger homeobox 2 ZFHX2 0.009 PREDICTED: zinc finger homeobox 2 LOC100290359 0.009 PREDICTED: hypothetical protein XP_002346414 ZFHX2 0.009 PREDICTED: zinc finger homeobox 2 LOC100287202 0.009 PREDICTED: hypothetical protein XP_002342114 NTN5 0.009 netrin 5 FAM171A2 0.009 family with sequence similarity 171, member A2 [Hom...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
