
| Name: LOC100293036 | Sequence: fasta or formatted (196aa) | NCBI GI: 239757559 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
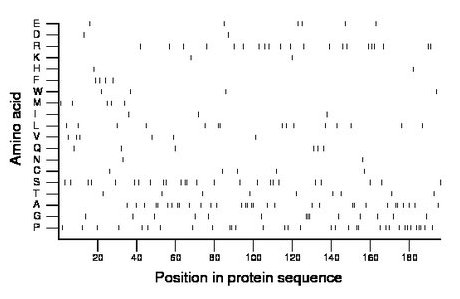
Amino acid Percentage Count Longest homopolymer A alanine 16.3 32 2 C cysteine 2.6 5 1 D aspartate 1.0 2 1 E glutamate 3.1 6 1 F phenylalanine 2.0 4 1 G glycine 7.1 14 3 H histidine 1.0 2 1 I isoleucine 1.5 3 1 K lysine 1.0 2 1 L leucine 7.7 15 2 M methionine 2.6 5 1 N asparagine 1.0 2 1 P proline 16.8 33 3 Q glutamine 3.1 6 1 R arginine 10.7 21 2 S serine 12.8 25 2 T threonine 4.6 9 1 V valine 3.1 6 1 W tryptophan 2.0 4 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100289065 0.879 PREDICTED: hypothetical protein LOC100289065 0.879 PREDICTED: hypothetical protein XP_002343789 SAMD1 0.090 sterile alpha motif domain containing 1 CIC 0.069 capicua homolog FLJ22184 0.063 PREDICTED: hypothetical protein FLJ22184 MAP7D1 0.063 MAP7 domain containing 1 LOC440829 0.058 PREDICTED: transmembrane protein 46-like HCN4 0.053 hyperpolarization activated cyclic nucleotide-gated p... WASL 0.053 Wiskott-Aldrich syndrome gene-like protein GLTSCR1 0.053 glioma tumor suppressor candidate region gene 1 [Ho... NACA 0.053 nascent polypeptide-associated complex alpha subuni... MDC1 0.053 mediator of DNA-damage checkpoint 1 LOC100289949 0.053 PREDICTED: hypothetical protein XP_002346877 LOC100290372 0.050 PREDICTED: hypothetical protein XP_002346788 LOC100288606 0.050 PREDICTED: hypothetical protein XP_002342652 IER5L 0.050 immediate early response 5-like WNK2 0.050 WNK lysine deficient protein kinase 2 UBE4B 0.050 ubiquitination factor E4B isoform 1 LOC100131102 0.047 PREDICTED: hypothetical protein LOC100293375 0.047 PREDICTED: hypothetical protein MUC7 0.045 mucin 7, secreted precursor FLJ22184 0.045 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.045 PREDICTED: hypothetical protein LOC80164 MUC7 0.045 mucin 7, secreted precursor MUC7 0.045 mucin 7, secreted precursor LOC100133756 0.045 PREDICTED: hypothetical protein, partial AEBP1 0.042 adipocyte enhancer binding protein 1 precursor [Homo... LOC100291634 0.042 PREDICTED: hypothetical protein XP_002346062 LOC100291096 0.042 PREDICTED: hypothetical protein XP_002346952Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
