
| Name: LOC100289949 | Sequence: fasta or formatted (326aa) | NCBI GI: 239748995 | |
|
Description: PREDICTED: hypothetical protein XP_002346877
| Not currently referenced in the text | ||
|
Composition:
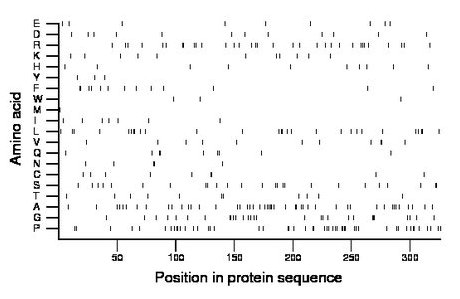
Amino acid Percentage Count Longest homopolymer A alanine 14.4 47 3 C cysteine 2.8 9 2 D aspartate 3.7 12 1 E glutamate 2.8 9 1 F phenylalanine 4.0 13 2 G glycine 8.9 29 3 H histidine 3.1 10 1 I isoleucine 2.1 7 1 K lysine 3.4 11 1 L leucine 8.6 28 2 M methionine 0.3 1 1 N asparagine 1.5 5 2 P proline 15.3 50 4 Q glutamine 2.8 9 2 R arginine 10.1 33 2 S serine 7.7 25 2 T threonine 3.4 11 1 V valine 3.4 11 2 W tryptophan 0.9 3 1 Y tyrosine 0.9 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346877 TAF4 0.053 TBP-associated factor 4 SYNJ1 0.038 synaptojanin 1 isoform c SYNJ1 0.038 synaptojanin 1 isoform a SYNJ1 0.038 synaptojanin 1 isoform b SPEG 0.038 SPEG complex locus SYNJ1 0.037 synaptojanin 1 isoform d FBRS 0.037 fibrosin SAMD1 0.037 sterile alpha motif domain containing 1 FLJ22184 0.033 PREDICTED: hypothetical protein FLJ22184 MAST1 0.033 microtubule associated serine/threonine kinase 1 [Ho... FLJ22184 0.033 PREDICTED: hypothetical protein FLJ22184 RUSC2 0.033 RUN and SH3 domain containing 2 RUSC2 0.033 RUN and SH3 domain containing 2 LOC731282 0.033 PREDICTED: hypothetical protein SH3BP1 0.033 SH3-domain binding protein 1 LOC729370 0.030 PREDICTED: hypothetical protein SFPQ 0.030 splicing factor proline/glutamine rich (polypyrimidin... LOC100291743 0.030 PREDICTED: hypothetical protein XP_002344931 LOC100289969 0.030 PREDICTED: hypothetical protein XP_002347692 LOC100288859 0.030 PREDICTED: hypothetical protein XP_002343508 DIAPH1 0.030 diaphanous 1 isoform 2 DIAPH1 0.030 diaphanous 1 isoform 1 ENAH 0.030 enabled homolog isoform a ENAH 0.030 enabled homolog isoform b LOC100293036 0.030 PREDICTED: hypothetical protein LOC100292122 0.030 PREDICTED: hypothetical protein XP_002345138 LOC100286986 0.030 PREDICTED: hypothetical protein XP_002344191 BSN 0.030 bassoon protein SYN1 0.030 synapsin I isoform IaHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
