
| Name: LOC100292842 | Sequence: fasta or formatted (121aa) | NCBI GI: 239757051 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
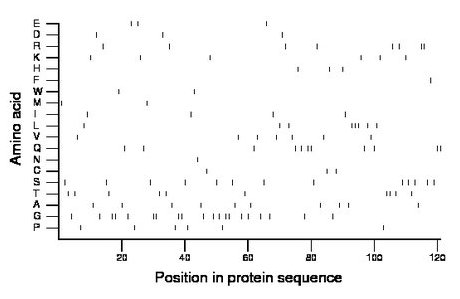
Amino acid Percentage Count Longest homopolymer A alanine 8.3 10 1 C cysteine 2.5 3 1 D aspartate 2.5 3 1 E glutamate 2.5 3 1 F phenylalanine 0.8 1 1 G glycine 16.5 20 2 H histidine 2.5 3 1 I isoleucine 3.3 4 1 K lysine 5.0 6 1 L leucine 6.6 8 3 M methionine 1.7 2 1 N asparagine 0.8 1 1 P proline 5.0 6 1 Q glutamine 9.1 11 2 R arginine 6.6 8 2 S serine 10.7 13 1 T threonine 8.3 10 2 V valine 5.8 7 1 W tryptophan 1.7 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291315 1.000 PREDICTED: hypothetical protein XP_002347903 LOC100287130 1.000 PREDICTED: hypothetical protein XP_002343640 EIF3H 0.031 eukaryotic translation initiation factor 3, subunit 3... LOC100132345 0.031 PREDICTED: hypothetical protein MAF 0.031 v-maf musculoaponeurotic fibrosarcoma oncogene homol... MAF 0.031 v-maf musculoaponeurotic fibrosarcoma oncogene homolo... LOC100293268 0.027 PREDICTED: hypothetical protein KHSRP 0.027 KH-type splicing regulatory protein RING1 0.027 ring finger protein 1 DDX51 0.027 DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 UPF1 0.027 regulator of nonsense transcripts 1 HNRNPA1 0.022 heterogeneous nuclear ribonucleoprotein A1 isoform b... SHANK2 0.022 SH3 and multiple ankyrin repeat domains 2 isoform 1... BHLHE22 0.022 basic helix-loop-helix domain containing, class B, 5... FGA 0.022 fibrinogen, alpha polypeptide isoform alpha prepropr... FGA 0.022 fibrinogen, alpha polypeptide isoform alpha-E preprop... GSK3A 0.018 glycogen synthase kinase 3 alpha STOX2 0.018 storkhead box 2 CASC3 0.018 metastatic lymph node 51 FIZ1 0.018 FLT3-interacting zinc finger 1 TRIM67 0.018 tripartite motif-containing 67 EWSR1 0.013 Ewing sarcoma breakpoint region 1 isoform 5 AGAP2 0.013 centaurin, gamma 1 isoform PIKE-L C6orf174 0.013 hypothetical protein LOC387104 LOC100288382 0.013 PREDICTED: hypothetical protein XP_002342213 LOR 0.013 loricrin KRT9 0.013 keratin 9 RYR1 0.013 skeletal muscle ryanodine receptor isoform 2 RYR1 0.013 skeletal muscle ryanodine receptor isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
