
| Name: LOC100291394 | Sequence: fasta or formatted (103aa) | NCBI GI: 239748601 | |
|
Description: PREDICTED: hypothetical protein XP_002346737
| Not currently referenced in the text | ||
|
Composition:
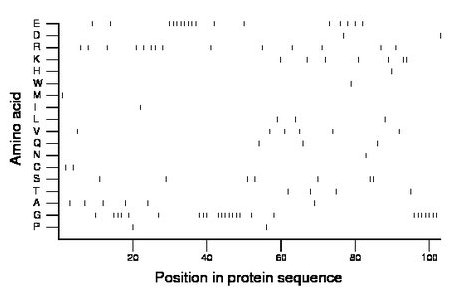
Amino acid Percentage Count Longest homopolymer A alanine 5.8 6 1 C cysteine 1.9 2 1 D aspartate 1.9 2 1 E glutamate 16.5 17 8 F phenylalanine 0.0 0 0 G glycine 24.3 25 7 H histidine 1.0 1 1 I isoleucine 1.0 1 1 K lysine 6.8 7 2 L leucine 2.9 3 1 M methionine 1.0 1 1 N asparagine 1.0 1 1 P proline 1.9 2 1 Q glutamine 2.9 3 1 R arginine 13.6 14 2 S serine 6.8 7 2 T threonine 3.9 4 1 V valine 5.8 6 1 W tryptophan 1.0 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346737 LOC100291586 1.000 PREDICTED: hypothetical protein XP_002345859 LOC100288032 1.000 PREDICTED: hypothetical protein XP_002342573 DHX57 0.096 DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 AEBP2 0.080 AE binding protein 2 isoform b AEBP2 0.080 AE binding protein 2 isoform a FAM9A 0.080 family with sequence similarity 9, member A PURA 0.069 purine-rich element binding protein A RPGR 0.064 retinitis pigmentosa GTPase regulator isoform C [Hom... XYLT1 0.064 xylosyltransferase I LTK 0.059 leukocyte receptor tyrosine kinase isoform 1 precurs... ANO8 0.053 anoctamin 8 NPAS3 0.053 neuronal PAS domain protein 3 isoform 2 NPAS3 0.053 neuronal PAS domain protein 3 isoform 1 FOXD1 0.053 forkhead box D1 FMN2 0.053 formin 2 RSBN1L 0.053 round spermatid basic protein 1-like DMWD 0.053 dystrophia myotonica-containing WD repeat motif prot... AR 0.053 androgen receptor isoform 1 YY1 0.053 YY1 transcription factor GIGYF1 0.053 PERQ amino acid rich, with GYF domain 1 CACNG8 0.053 voltage-dependent calcium channel gamma-8 subunit [H... LOC100287290 0.048 PREDICTED: hypothetical protein LOC100287290 0.048 PREDICTED: hypothetical protein XP_002342446 TUSC1 0.048 tumor suppressor candidate 1 EHMT1 0.048 euchromatic histone-lysine N-methyltransferase 1 is... EHMT1 0.048 euchromatic histone-lysine N-methyltransferase 1 is... MNX1 0.048 homeo box HB9 LOC100288007 0.048 PREDICTED: hypothetical protein LOC100288007 0.048 PREDICTED: hypothetical protein XP_002342192Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
