
| Name: LOC100289342 | Sequence: fasta or formatted (110aa) | NCBI GI: 239743644 | |
|
Description: PREDICTED: hypothetical protein XP_002342912
| Not currently referenced in the text | ||
|
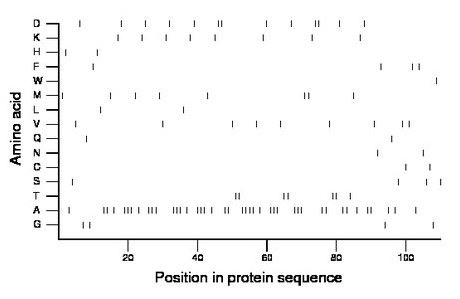
Composition:
Amino acid Percentage Count Longest homopolymer A alanine 38.2 42 4 C cysteine 1.8 2 1 D aspartate 11.8 13 2 E glutamate 0.0 0 0 F phenylalanine 3.6 4 1 G glycine 3.6 4 1 H histidine 1.8 2 1 I isoleucine 0.0 0 0 K lysine 7.3 8 1 L leucine 1.8 2 1 M methionine 7.3 8 2 N asparagine 1.8 2 1 P proline 0.0 0 0 Q glutamine 1.8 2 1 R arginine 0.0 0 0 S serine 3.6 4 1 T threonine 6.4 7 2 V valine 8.2 9 1 W tryptophan 0.9 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342912 LOC727872 0.882 PREDICTED: hypothetical protein AMOT 0.054 angiomotin isoform 2 AMOT 0.054 angiomotin isoform 1 FAM48B1 0.054 hypothetical protein LOC100130302 LOC100289830 0.048 PREDICTED: hypothetical protein XP_002347519 MAP4 0.048 microtubule-associated protein 4 isoform 4 MAP4 0.048 microtubule-associated protein 4 isoform 1 MAZ 0.038 MYC-associated zinc finger protein isoform 2 MAZ 0.038 MYC-associated zinc finger protein isoform 1 LOC342346 0.032 hypothetical protein LOC342346 RCOR1 0.032 REST corepressor 1 HOXA13 0.032 homeobox A13 FOXD1 0.027 forkhead box D1 HCFC1 0.027 host cell factor 1 MARCKS 0.027 myristoylated alanine-rich protein kinase C substra... COL15A1 0.027 alpha 1 type XV collagen precursor TLN2 0.027 talin 2 SP8 0.016 Sp8 transcription factor isoform 1 SP8 0.016 Sp8 transcription factor isoform 2 LOC100130667 0.016 PREDICTED: hypothetical protein LOC100130667 0.016 PREDICTED: hypothetical protein LOC100130667 0.016 PREDICTED: hypothetical protein MAP1A 0.016 microtubule-associated protein 1A TTN 0.016 titin isoform novex-3 TTN 0.016 titin isoform N2-A TTN 0.016 titin isoform novex-2 TTN 0.016 titin isoform novex-1 TTN 0.016 titin isoform N2-B LBXCOR1 0.016 LBXCOR1 homologHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
