
| Name: LOC100289830 | Sequence: fasta or formatted (426aa) | NCBI GI: 239750735 | |
|
Description: PREDICTED: hypothetical protein XP_002347519
| Not currently referenced in the text | ||
|
Composition:
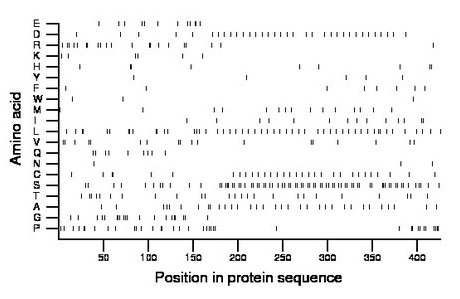
Amino acid Percentage Count Longest homopolymer A alanine 7.7 33 1 C cysteine 7.3 31 2 D aspartate 8.0 34 1 E glutamate 2.8 12 2 F phenylalanine 1.9 8 1 G glycine 4.7 20 2 H histidine 2.1 9 2 I isoleucine 3.3 14 1 K lysine 1.4 6 1 L leucine 12.0 51 2 M methionine 3.5 15 1 N asparagine 0.7 3 1 P proline 8.5 36 2 Q glutamine 2.1 9 1 R arginine 4.9 21 2 S serine 17.1 73 3 T threonine 5.9 25 1 V valine 4.2 18 1 W tryptophan 0.7 3 1 Y tyrosine 1.2 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347519 LOC100288531 0.102 PREDICTED: hypothetical protein XP_002343337 LOC727872 0.026 PREDICTED: hypothetical protein LOC729815 0.022 PREDICTED: hypothetical protein LOC729815 0.022 PREDICTED: hypothetical protein LOC100291627 0.014 PREDICTED: hypothetical protein XP_002345251 LOC100289867 0.014 PREDICTED: hypothetical protein XP_002348034 GRN 0.013 granulin precursor TNRC18 0.013 trinucleotide repeat containing 18 SPEG 0.013 SPEG complex locus NLRP4 0.012 NLR family, pyrin domain containing 4 MDFI 0.012 MyoD family inhibitor CHD9 0.012 chromodomain helicase DNA binding protein 9 LOC100293112 0.012 PREDICTED: hypothetical protein LOC100131251 0.012 PREDICTED: hypothetical protein LOC100288817 0.012 PREDICTED: hypothetical protein LOC100131251 0.012 PREDICTED: hypothetical protein LOC100288817 0.012 PREDICTED: hypothetical protein XP_002343307 LOC100289342 0.011 PREDICTED: hypothetical protein XP_002342912 KRTAP5-9 0.011 keratin associated protein 5-9 MYST3 0.011 MYST histone acetyltransferase (monocytic leukemia)... MYST3 0.011 MYST histone acetyltransferase (monocytic leukemia)... MYST3 0.011 MYST histone acetyltransferase (monocytic leukemia)... FBXL20 0.011 F-box and leucine-rich repeat protein 20 APC2 0.009 adenomatosis polyposis coli 2 NRG1 0.009 neuregulin 1 isoform GGF2 INPPL1 0.009 inositol polyphosphate phosphatase-like 1 PODXL 0.008 podocalyxin-like isoform 2 precursor PODXL 0.008 podocalyxin-like isoform 1 precursor AHNAK2 0.008 AHNAK nucleoprotein 2Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
