
| Name: LOC100288705 | Sequence: fasta or formatted (79aa) | NCBI GI: 239741589 | |
|
Description: PREDICTED: hypothetical protein XP_002342258
| Not currently referenced in the text | ||
|
Composition:
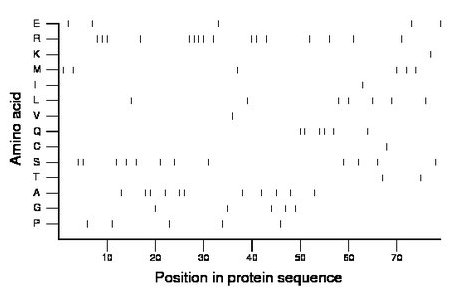
Amino acid Percentage Count Longest homopolymer A alanine 13.9 11 2 C cysteine 1.3 1 1 D aspartate 0.0 0 0 E glutamate 6.3 5 1 F phenylalanine 0.0 0 0 G glycine 6.3 5 1 H histidine 0.0 0 0 I isoleucine 1.3 1 1 K lysine 1.3 1 1 L leucine 8.9 7 1 M methionine 7.6 6 1 N asparagine 0.0 0 0 P proline 6.3 5 1 Q glutamine 7.6 6 2 R arginine 20.3 16 4 S serine 15.2 12 2 T threonine 2.5 2 1 V valine 1.3 1 1 W tryptophan 0.0 0 0 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342258 LOC100292994 1.000 PREDICTED: hypothetical protein LOC100291003 1.000 PREDICTED: hypothetical protein XP_002348235 DEDD2 0.079 death effector domain-containing DNA binding protei... LOC645321 0.056 PREDICTED: hypothetical protein LOC100293818 0.048 PREDICTED: hypothetical protein LOC100128949 0.040 PREDICTED: hypothetical protein CACNA1B 0.040 calcium channel, voltage-dependent, N type, alpha 1B ... LOC339742 0.040 PREDICTED: hypothetical protein LOC100293212 0.040 PREDICTED: hypothetical protein LOC100289796 0.040 PREDICTED: hypothetical protein XP_002346485 LOC100288812 0.040 PREDICTED: hypothetical protein XP_002342329 SORBS1 0.040 sorbin and SH3 domain containing 1 isoform 4 SORBS1 0.040 sorbin and SH3 domain containing 1 isoform 3 INPP5J 0.040 phosphatidylinositol (4,5) bisphosphate 5-phosphatas... SYNCRIP 0.032 synaptotagmin binding, cytoplasmic RNA interacting ... SYNCRIP 0.032 synaptotagmin binding, cytoplasmic RNA interacting ... SYNCRIP 0.032 synaptotagmin binding, cytoplasmic RNA interacting ... LOC100128949 0.032 PREDICTED: hypothetical protein LOC100128949 0.032 PREDICTED: hypothetical protein SALL3 0.032 sal-like 3 SYNCRIP 0.032 synaptotagmin binding, cytoplasmic RNA interacting ... SYNCRIP 0.032 synaptotagmin binding, cytoplasmic RNA interacting ... SYNCRIP 0.032 synaptotagmin binding, cytoplasmic RNA interacting ... LOC729580 0.032 PREDICTED: hypothetical protein LOC729580 0.032 PREDICTED: hypothetical protein LOC729580 0.032 PREDICTED: hypothetical protein LOC388279 0.032 PREDICTED: hypothetical protein APC2 0.032 adenomatosis polyposis coli 2 SRRM2 0.032 splicing coactivator subunit SRm300Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
