
| Name: LOC728449 | Sequence: fasta or formatted (199aa) | NCBI GI: 113421471 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
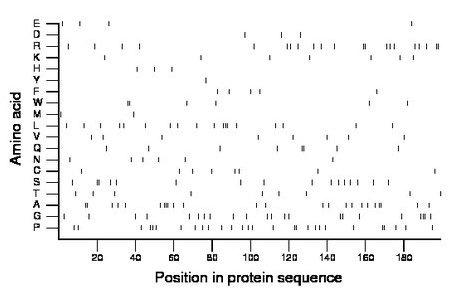
Amino acid Percentage Count Longest homopolymer A alanine 12.1 24 3 C cysteine 4.0 8 1 D aspartate 1.5 3 1 E glutamate 2.0 4 1 F phenylalanine 2.5 5 1 G glycine 11.1 22 3 H histidine 1.5 3 1 I isoleucine 0.0 0 0 K lysine 3.5 7 1 L leucine 9.0 18 3 M methionine 1.0 2 1 N asparagine 3.0 6 1 P proline 14.1 28 2 Q glutamine 4.0 8 2 R arginine 10.6 21 2 S serine 8.0 16 2 T threonine 4.5 9 1 V valine 4.0 8 1 W tryptophan 3.0 6 2 Y tyrosine 0.5 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100132646 0.792 PREDICTED: hypothetical protein LOC100127930 0.030 PREDICTED: hypothetical protein RBMX 0.027 RNA binding motif protein, X-linked FOXE1 0.025 forkhead box E1 LOC100292745 0.025 PREDICTED: hypothetical protein LOC100290074 0.025 PREDICTED: hypothetical protein XP_002346855 LOC100287034 0.025 PREDICTED: hypothetical protein XP_002342693 RERE 0.025 atrophin-1 like protein isoform b RERE 0.025 atrophin-1 like protein isoform a RERE 0.025 atrophin-1 like protein isoform a FLJ37078 0.022 hypothetical protein LOC222183 TCERG1L 0.022 transcription elongation regulator 1-like PTCRA 0.020 pre T-cell antigen receptor alpha LOC100288496 0.020 PREDICTED: hypothetical protein XP_002342747 LOC728597 0.017 PREDICTED: doublecortin domain-containing protein E... LOC728597 0.017 PREDICTED: doublecortin domain-containing protein E... GPR153 0.017 G protein-coupled receptor 153 HOXD11 0.017 homeobox D11 LOC100294010 0.017 PREDICTED: hypothetical protein LOC100293596 0.017 PREDICTED: similar to mucin LOC100291514 0.017 PREDICTED: hypothetical protein XP_002347163 LOC100289311 0.017 PREDICTED: hypothetical protein XP_002343026 GLI4 0.017 GLI-Kruppel family member GLI4 TMEM181 0.017 G protein-coupled receptor 178 ZNF574 0.017 zinc finger protein 574 C1orf53 0.017 hypothetical protein LOC388722 LOC100291756 0.017 PREDICTED: hypothetical protein XP_002345525 LOC100130465 0.017 PREDICTED: hypothetical protein LOC100293375 0.017 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
