
| Name: SUFU | Sequence: fasta or formatted (484aa) | NCBI GI: 19923479 | |
|
Description: suppressor of fused
|
Referenced in:
| ||
|
Composition:
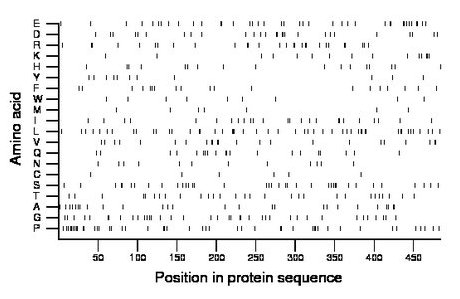
Amino acid Percentage Count Longest homopolymer A alanine 5.4 26 2 C cysteine 1.2 6 1 D aspartate 5.6 27 2 E glutamate 8.1 39 2 F phenylalanine 3.9 19 1 G glycine 7.9 38 2 H histidine 3.5 17 1 I isoleucine 5.2 25 2 K lysine 3.1 15 2 L leucine 10.1 49 2 M methionine 1.7 8 1 N asparagine 2.5 12 1 P proline 9.5 46 2 Q glutamine 4.1 20 2 R arginine 5.8 28 2 S serine 7.6 37 2 T threonine 6.0 29 1 V valine 4.8 23 2 W tryptophan 1.9 9 1 Y tyrosine 2.3 11 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 suppressor of fused AP4E1 0.013 adaptor-related protein complex 4, epsilon 1 subunit... TBX21 0.012 T-box 21 LOC100291760 0.012 PREDICTED: hypothetical protein XP_002344912 LOC100290674 0.012 PREDICTED: hypothetical protein XP_002347612 LOC100288732 0.012 PREDICTED: hypothetical protein XP_002343433 OTUD1 0.010 OTU domain containing 1 ZFPM1 0.010 zinc finger protein, multitype 1 COL14A1 0.010 collagen, type XIV, alpha 1 COL4A5 0.009 type IV collagen alpha 5 isoform 2 precursor COL4A5 0.009 type IV collagen alpha 5 isoform 1 precursor COL4A5 0.009 type IV collagen alpha 5 isoform 3 precursor COL17A1 0.009 alpha 1 type XVII collagen TMEM158 0.009 Ras-induced senescence 1 COLEC12 0.009 collectin sub-family member 12 CPSF6 0.008 cleavage and polyadenylation specific factor 6, 68 ... KRTAP4-6 0.008 PREDICTED: keratin associated protein 4.6 FASTK 0.008 Fas-activated serine/threonine kinase isoform 4 [Hom... EMID2 0.008 EMI domain containing 2 COL15A1 0.008 alpha 1 type XV collagen precursor VASP 0.007 vasodilator-stimulated phosphoprotein C21orf70 0.007 hypothetical protein LOC85395 LOC100133554 0.007 PREDICTED: hypothetical protein LOC100133554 0.007 PREDICTED: hypothetical protein TNRC18 0.007 trinucleotide repeat containing 18 LOC100133142 0.007 PREDICTED: similar to zinc finger protein 208 [Homo... LOC100133142 0.007 PREDICTED: similar to zinc finger protein 208 [Homo... PCDH15 0.007 protocadherin 15 isoform CD1-4 precursor PCDH15 0.007 protocadherin 15 isoform CD1-10 precursor PCDH15 0.007 protocadherin 15 isoform CD1-9 precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
