
| Name: SKIV2L2 | Sequence: fasta or formatted (1042aa) | NCBI GI: 193211480 | |
|
Description: superkiller viralicidic activity 2-like 2
|
Referenced in: DEAD / H Helicase Family
| ||
|
Composition:
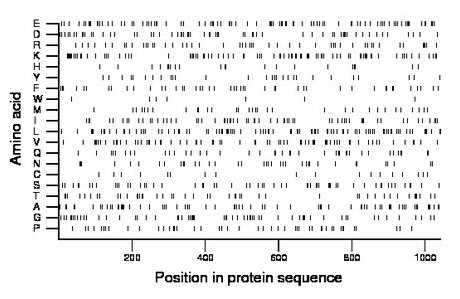
Amino acid Percentage Count Longest homopolymer A alanine 6.9 72 2 C cysteine 1.7 18 1 D aspartate 5.9 61 2 E glutamate 8.6 90 2 F phenylalanine 4.4 46 1 G glycine 5.9 61 2 H histidine 2.0 21 2 I isoleucine 6.1 64 2 K lysine 8.4 88 2 L leucine 9.1 95 3 M methionine 3.3 34 2 N asparagine 3.6 38 2 P proline 4.4 46 2 Q glutamine 3.8 40 2 R arginine 5.0 52 2 S serine 5.7 59 2 T threonine 4.4 46 2 V valine 7.2 75 3 W tryptophan 0.7 7 1 Y tyrosine 2.8 29 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 superkiller viralicidic activity 2-like 2 SKIV2L 0.267 superkiller viralicidic activity 2-like homolog [Hom... HFM1 0.043 HFM1 protein ASCC3 0.042 activating signal cointegrator 1 complex subunit 3 i... POLQ 0.040 DNA polymerase theta SNRNP200 0.035 activating signal cointegrator 1 complex subunit 3-l... LOC652147 0.033 PREDICTED: similar to U5 snRNP-specific 200kD prote... HELQ 0.030 DNA helicase HEL308 DDX60L 0.029 DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like [Hom... DDX60 0.027 DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 RECQL 0.008 RecQ protein-like RECQL 0.008 RecQ protein-like DDX54 0.007 DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 isoform 2 ... DDX54 0.007 DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 isoform 1... SUPV3L1 0.007 suppressor of var1, 3-like 1 DDX50 0.005 nucleolar protein GU2 DHX36 0.005 DEAH (Asp-Glu-Ala-His) box polypeptide 36 isoform 2... DHX36 0.005 DEAH (Asp-Glu-Ala-His) box polypeptide 36 isoform 1... MYO5B 0.005 myosin VB RECQL4 0.004 RecQ protein-like 4 WRN 0.004 Werner syndrome protein LOC652522 0.004 PREDICTED: similar to Werner syndrome ATP-dependent... DDX27 0.004 DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 MYO5A 0.004 myosin VA isoform 2 MYO5A 0.004 myosin VA isoform 1 C14orf106 0.003 chromosome 14 open reading frame 106 FCRL4 0.003 Fc receptor-like 4 precursor PTGES2 0.003 prostaglandin E synthase 2 CHD4 0.003 chromodomain helicase DNA binding protein 4 MYO5C 0.003 myosin VCHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
