
| Name: C9orf140 | Sequence: fasta or formatted (394aa) | NCBI GI: 190886454 | |
|
Description: tumor specificity and mitosis phase-dependent expression protein
| Not currently referenced in the text | ||
|
Composition:
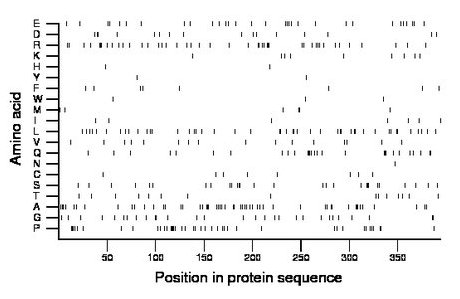
Amino acid Percentage Count Longest homopolymer A alanine 13.5 53 3 C cysteine 2.0 8 1 D aspartate 4.1 16 2 E glutamate 7.9 31 1 F phenylalanine 2.0 8 1 G glycine 7.4 29 2 H histidine 0.5 2 1 I isoleucine 1.8 7 1 K lysine 2.3 9 1 L leucine 11.7 46 2 M methionine 1.5 6 2 N asparagine 0.3 1 1 P proline 10.4 41 5 Q glutamine 7.1 28 3 R arginine 11.4 45 3 S serine 7.1 28 3 T threonine 3.8 15 1 V valine 4.1 16 1 W tryptophan 0.8 3 1 Y tyrosine 0.5 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 tumor specificity and mitosis phase-dependent expre... ZNF716 0.384 PREDICTED: zinc finger protein 716 C6orf26 0.034 hypothetical protein LOC401251 KIAA0754 0.030 hypothetical protein LOC643314 TBX2 0.026 T-box 2 MAP7 0.026 microtubule-associated protein 7 C19orf61 0.025 hypothetical protein LOC56006 SAMD1 0.025 sterile alpha motif domain containing 1 CDKN1C 0.025 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.025 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.025 cyclin-dependent kinase inhibitor 1C isoform a MAP7D1 0.025 MAP7 domain containing 1 WNK2 0.024 WNK lysine deficient protein kinase 2 LOC645321 0.024 PREDICTED: hypothetical protein LOC645321 0.024 PREDICTED: hypothetical protein KDM6B 0.024 lysine (K)-specific demethylase 6B DACT3 0.024 thymus expressed gene 3-like NES 0.022 nestin RFX1 0.022 regulatory factor X1 MARCH11 0.022 membrane-associated ring finger (C3HC4) 11 C17orf96 0.021 hypothetical protein LOC100170841 PRR12 0.021 proline rich 12 MARCKS 0.020 myristoylated alanine-rich protein kinase C substra... TMEM88B 0.020 PREDICTED: hypothetical protein LOC643965 TMEM88B 0.020 PREDICTED: hypothetical protein LOC643965 CC2D1A 0.020 coiled-coil and C2 domain containing 1A NHS 0.020 Nance-Horan syndrome protein isoform 1 BZRAP1 0.020 peripheral benzodiazepine receptor-associated prote... BZRAP1 0.020 peripheral benzodiazepine receptor-associated prote... GGN 0.020 gametogenetinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
