
| Name: LOC100132826 | Sequence: fasta or formatted (234aa) | NCBI GI: 169210116 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
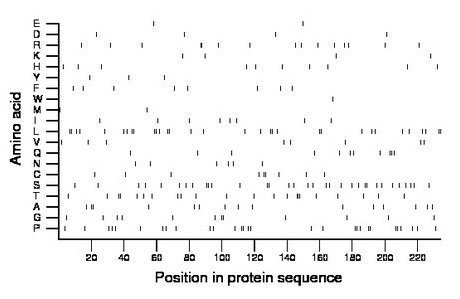
Amino acid Percentage Count Longest homopolymer A alanine 6.4 15 2 C cysteine 3.8 9 2 D aspartate 1.7 4 1 E glutamate 0.9 2 1 F phenylalanine 3.4 8 1 G glycine 5.1 12 2 H histidine 4.3 10 1 I isoleucine 3.4 8 1 K lysine 1.7 4 1 L leucine 13.7 32 2 M methionine 0.9 2 1 N asparagine 2.6 6 1 P proline 12.4 29 2 Q glutamine 4.7 11 2 R arginine 6.4 15 2 S serine 14.1 33 2 T threonine 9.4 22 3 V valine 3.4 8 1 W tryptophan 0.4 1 1 Y tyrosine 1.3 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC728836 0.982 PREDICTED: hypothetical protein LOC100132632 0.947 PREDICTED: hypothetical protein POM121L2 0.022 POM121 membrane glycoprotein-like 2 FLJ22184 0.020 PREDICTED: hypothetical protein FLJ22184 SHANK1 0.020 SH3 and multiple ankyrin repeat domains 1 KCNC3 0.018 Shaw-related voltage-gated potassium channel protein... LOC100292374 0.018 PREDICTED: similar to transmembrane protein 125 [Ho... LOC100290008 0.018 PREDICTED: hypothetical protein XP_002346440 LOC100288382 0.018 PREDICTED: hypothetical protein XP_002342213 LOC100288314 0.018 PREDICTED: hypothetical protein XP_002342147 LOC100292845 0.015 PREDICTED: hypothetical protein LOC100289230 0.015 PREDICTED: hypothetical protein XP_002342600 PODXL 0.015 podocalyxin-like isoform 1 precursor PDE4A 0.015 phosphodiesterase 4A isoform 3 PDE4A 0.015 phosphodiesterase 4A isoform 2 PDE4A 0.015 phosphodiesterase 4A isoform 1 MUC17 0.015 mucin 17 RCOR2 0.015 REST corepressor 2 BAT2 0.015 HLA-B associated transcript-2 PPRC1 0.015 peroxisome proliferator-activated receptor gamma, co... LMTK3 0.015 lemur tyrosine kinase 3 ULK2 0.013 unc-51-like kinase 2 ULK2 0.013 unc-51-like kinase 2 PHC3 0.013 polyhomeotic like 3 WASF1 0.013 Wiskott-Aldrich syndrome protein family member 1 [Ho... WASF1 0.013 Wiskott-Aldrich syndrome protein family member 1 [Ho... WASF1 0.013 Wiskott-Aldrich syndrome protein family member 1 [Ho... WASF1 0.013 Wiskott-Aldrich syndrome protein family member 1 [Hom... LOC100294448 0.013 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
