
| Name: LOC100134230 | Sequence: fasta or formatted (109aa) | NCBI GI: 169161476 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
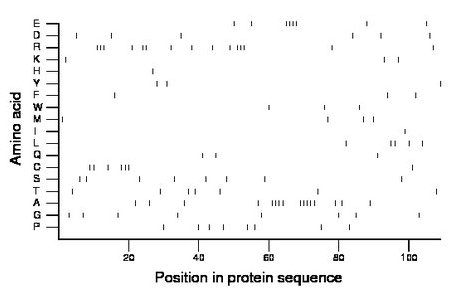
Amino acid Percentage Count Longest homopolymer A alanine 14.7 16 5 C cysteine 6.4 7 3 D aspartate 5.5 6 1 E glutamate 7.3 8 4 F phenylalanine 2.8 3 1 G glycine 7.3 8 1 H histidine 0.9 1 1 I isoleucine 0.9 1 1 K lysine 2.8 3 1 L leucine 4.6 5 2 M methionine 3.7 4 1 N asparagine 0.0 0 0 P proline 7.3 8 1 Q glutamine 2.8 3 1 R arginine 13.8 15 3 S serine 7.3 8 1 T threonine 6.4 7 1 V valine 0.0 0 0 W tryptophan 2.8 3 1 Y tyrosine 2.8 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein PDE4DIP 0.697 phosphodiesterase 4D interacting protein isoform 3 [... LOC100133167 0.057 PREDICTED: hypothetical protein LOC100133167 0.057 PREDICTED: hypothetical protein LOC100133167 0.057 PREDICTED: hypothetical protein HRH2 0.052 histamine receptor H2 isoform 1 MAP1A 0.028 microtubule-associated protein 1A PCNT 0.028 pericentrin EMILIN2 0.024 elastin microfibril interfacer 2 KIAA0368 0.024 KIAA0368 protein CASZ1 0.024 castor homolog 1, zinc finger isoform a CASZ1 0.024 castor homolog 1, zinc finger isoform b HOXB5 0.024 homeobox B5 PLEKHG4B 0.019 pleckstrin homology domain containing, family G (wi... DMRT3 0.019 doublesex and mab-3 related transcription factor 3 [... TTMA 0.019 hypothetical protein LOC645369 FLJ46210 0.019 hypothetical protein LOC389152 LOC100128025 0.014 PREDICTED: hypothetical protein LOC100128025 0.014 PREDICTED: hypothetical protein LOC100129203 0.014 PREDICTED: similar to mCG50504 LOC100128025 0.014 PREDICTED: hypothetical protein BAT3 0.014 HLA-B associated transcript-3 isoform b BAT3 0.014 HLA-B associated transcript-3 isoform b BAT3 0.014 HLA-B associated transcript-3 isoform b BAT3 0.014 HLA-B associated transcript-3 isoform a GLCCI1 0.014 glucocorticoid induced transcript 1 NRG3 0.014 neuregulin 3 LOC100291206 0.014 PREDICTED: hypothetical protein XP_002347014 KRTAP5-10 0.014 keratin associated protein 5-10 ACACB 0.014 acetyl-Coenzyme A carboxylase betaHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
