
| Name: LOC199800 | Sequence: fasta or formatted (153aa) | NCBI GI: 157415950 | |
|
Description: hypothetical protein LOC199800
| Not currently referenced in the text | ||
|
Composition:
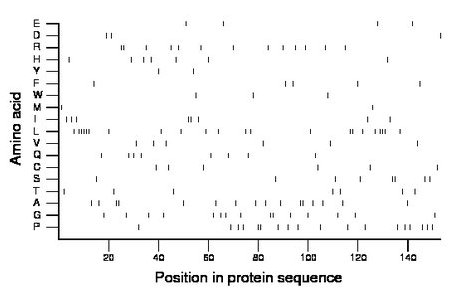
Amino acid Percentage Count Longest homopolymer A alanine 10.5 16 2 C cysteine 3.9 6 1 D aspartate 2.0 3 1 E glutamate 2.6 4 1 F phenylalanine 3.3 5 1 G glycine 9.8 15 2 H histidine 4.6 7 1 I isoleucine 5.2 8 2 K lysine 0.0 0 0 L leucine 14.4 22 5 M methionine 1.3 2 1 N asparagine 0.0 0 0 P proline 11.8 18 2 Q glutamine 5.2 8 1 R arginine 8.5 13 2 S serine 5.2 8 2 T threonine 4.6 7 1 V valine 3.9 6 1 W tryptophan 2.0 3 1 Y tyrosine 1.3 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC199800 LOC100294010 0.030 PREDICTED: hypothetical protein LOC100291514 0.030 PREDICTED: hypothetical protein XP_002347163 LOC100289311 0.030 PREDICTED: hypothetical protein XP_002343026 LOC100288801 0.027 PREDICTED: hypothetical protein XP_002342402 LOC100288239 0.023 PREDICTED: hypothetical protein XP_002343261 FOXN1 0.023 forkhead box N1 ATOH8 0.023 atonal homolog 8 LOC100289316 0.023 PREDICTED: hypothetical protein LOC729660 0.023 PREDICTED: similar to Putative uncharacterized prot... LOC100289316 0.023 PREDICTED: hypothetical protein XP_002343619 SRCAP 0.017 Snf2-related CBP activator protein POM121L12 0.017 POM121 membrane glycoprotein-like 12 LOC643355 0.017 PREDICTED: hypothetical protein LOC100289704 0.017 PREDICTED: hypothetical protein XP_002347777 LOC100287039 0.017 PREDICTED: hypothetical protein XP_002342225 LOC100130431 0.013 PREDICTED: hypothetical protein LOC100130431 0.013 PREDICTED: hypothetical protein LOC100130431 0.013 PREDICTED: hypothetical protein LOC646862 0.013 PREDICTED: hypothetical protein LOC646862 LOC100293296 0.013 PREDICTED: hypothetical protein LOC646862 0.013 PREDICTED: hypothetical protein LOC646862 LOC100290436 0.013 PREDICTED: hypothetical protein XP_002347616 LOC646862 0.013 PREDICTED: hypothetical protein LOC646862 LOC100287267 0.013 PREDICTED: hypothetical protein XP_002343446 C17orf68 0.013 alpha accessory factor 132 DUSP8 0.010 dual specificity phosphatase 8 SELV 0.010 selenoprotein V ZDHHC18 0.010 zinc finger, DHHC-type containing 18 DBP 0.010 D site of albumin promoter (albumin D-box) binding p...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
