
| Name: ZNF839 | Sequence: fasta or formatted (927aa) | NCBI GI: 153251840 | |
|
Description: zinc finger protein 839
| Not currently referenced in the text | ||
|
Composition:
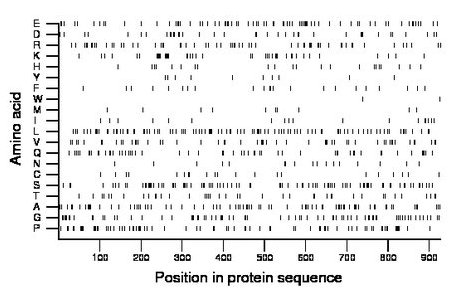
Amino acid Percentage Count Longest homopolymer A alanine 8.8 82 3 C cysteine 2.3 21 1 D aspartate 4.2 39 2 E glutamate 8.2 76 2 F phenylalanine 2.4 22 2 G glycine 8.6 80 6 H histidine 2.4 22 1 I isoleucine 1.9 18 1 K lysine 4.5 42 2 L leucine 10.8 100 2 M methionine 1.2 11 1 N asparagine 1.8 17 2 P proline 8.8 82 7 Q glutamine 5.9 55 2 R arginine 6.8 63 3 S serine 9.7 90 4 T threonine 4.5 42 3 V valine 5.7 53 2 W tryptophan 0.2 2 1 Y tyrosine 1.1 10 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 zinc finger protein 839 LOC100132864 0.117 PREDICTED: hypothetical protein LOC729162 0.066 PREDICTED: similar to hCG1981372 LOC729162 0.064 PREDICTED: similar to hCG1981372 LOC389906 0.064 PREDICTED: similar to Serine/threonine-protein kina... LOC389906 0.034 PREDICTED: similar to Serine/threonine-protein kina... LOC100293492 0.012 PREDICTED: hypothetical protein ZBTB7B 0.010 zinc finger and BTB domain containing 7B LOC100293130 0.010 PREDICTED: similar to kruppel-related zinc finger p... LOC100293130 0.010 PREDICTED: similar to kruppel-related zinc finger p... LOC100293130 0.010 PREDICTED: similar to kruppel-related zinc finger p... FBLIM1 0.010 filamin-binding LIM protein-1 isoform b FBLIM1 0.010 filamin-binding LIM protein-1 isoform a NACA 0.010 nascent polypeptide-associated complex alpha subuni... SOCS7 0.009 suppressor of cytokine signaling 7 TTN 0.009 titin isoform N2-A SFPQ 0.008 splicing factor proline/glutamine rich (polypyrimidin... FLJ22184 0.008 PREDICTED: hypothetical protein FLJ22184 HSF4 0.008 heat shock transcription factor 4 isoform a SRCAP 0.008 Snf2-related CBP activator protein CREBBP 0.008 CREB binding protein isoform b CREBBP 0.008 CREB binding protein isoform a IRS4 0.008 insulin receptor substrate 4 MEF2D 0.008 myocyte enhancer factor 2D MAPK8IP1 0.008 mitogen-activated protein kinase 8 interacting protei... YLPM1 0.008 YLP motif containing 1 CAPNS1 0.007 calpain, small subunit 1 CAPNS1 0.007 calpain, small subunit 1 HSPBP1 0.007 hsp70-interacting protein HSPBP1 0.007 hsp70-interacting proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
