
| Name: C11orf20 | Sequence: fasta or formatted (200aa) | NCBI GI: 148839350 | |
|
Description: hypothetical protein LOC25858
| Not currently referenced in the text | ||
|
Composition:
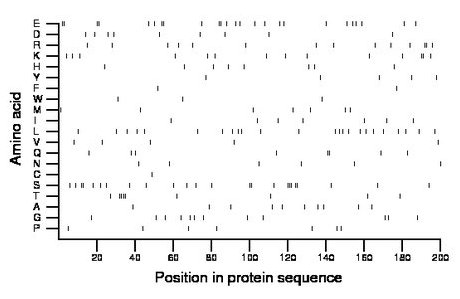
Amino acid Percentage Count Longest homopolymer A alanine 5.0 10 1 C cysteine 0.5 1 1 D aspartate 4.5 9 1 E glutamate 12.5 25 2 F phenylalanine 1.0 2 1 G glycine 6.0 12 1 H histidine 4.0 8 1 I isoleucine 3.5 7 1 K lysine 7.0 14 2 L leucine 12.0 24 1 M methionine 3.5 7 1 N asparagine 3.0 6 1 P proline 3.5 7 1 Q glutamine 4.0 8 2 R arginine 7.0 14 2 S serine 12.0 24 3 T threonine 4.5 9 4 V valine 2.5 5 1 W tryptophan 1.5 3 1 Y tyrosine 2.5 5 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC25858 PCTK1 0.013 PCTAIRE protein kinase 1 isoform 2 PCTK1 0.013 PCTAIRE protein kinase 1 isoform 1 EFCAB5 0.011 EF-hand calcium binding domain 5 isoform a CCDC108 0.011 coiled-coil domain containing 108 isoform 1 GTF2IRD2 0.011 GTF2I repeat domain containing 2 PITPNC1 0.011 phosphatidylinositol transfer protein, cytoplasmic 1... PITPNC1 0.011 phosphatidylinositol transfer protein, cytoplasmic 1... WNK2 0.008 WNK lysine deficient protein kinase 2 ANKRD12 0.008 ankyrin repeat domain 12 isoform 2 ANKRD12 0.008 ankyrin repeat domain 12 isoform 1 ARHGEF15 0.008 Rho guanine exchange factor 15 LOC100293874 0.008 PREDICTED: hypothetical protein WDR60 0.008 WD repeat domain 60 ARID4A 0.008 retinoblastoma-binding protein 1 isoform III ARID4A 0.008 retinoblastoma-binding protein 1 isoform II ARID4A 0.008 retinoblastoma-binding protein 1 isoform I HTT 0.005 huntingtin NGF 0.005 nerve growth factor, beta polypeptide precursor [Hom... LOC100293081 0.005 PREDICTED: hypothetical protein LOC100290319 0.005 PREDICTED: hypothetical protein XP_002347063 LOC100288748 0.005 PREDICTED: hypothetical protein XP_002342905Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
