
| Name: PLSCR5 | Sequence: fasta or formatted (271aa) | NCBI GI: 146229354 | |
|
Description: phospholipid scramblase family, member 5
|
Referenced in:
| ||
|
Composition:
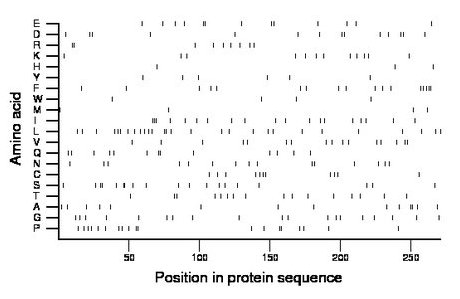
Amino acid Percentage Count Longest homopolymer A alanine 5.2 14 2 C cysteine 4.1 11 1 D aspartate 5.2 14 1 E glutamate 4.8 13 2 F phenylalanine 6.3 17 2 G glycine 7.0 19 1 H histidine 1.1 3 1 I isoleucine 7.0 19 3 K lysine 4.4 12 1 L leucine 10.3 28 2 M methionine 1.5 4 1 N asparagine 5.5 15 1 P proline 7.0 19 2 Q glutamine 4.8 13 2 R arginine 3.3 9 2 S serine 7.0 19 2 T threonine 5.9 16 1 V valine 5.9 16 1 W tryptophan 1.5 4 1 Y tyrosine 2.2 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 phospholipid scramblase family, member 5 PLSCR1 0.535 phospholipid scramblase 1 PLSCR2 0.507 phospholipid scramblase 2 PLSCR3 0.411 phospholipid scramblase 3 PLSCR4 0.338 phospholipid scramblase 4 isoform a PLSCR4 0.338 phospholipid scramblase 4 isoform a PLSCR4 0.338 phospholipid scramblase 4 isoform a PLSCR4 0.177 phospholipid scramblase 4 isoform b LOC440981 0.117 PREDICTED: similar to hCG1794790 LOC440981 0.112 PREDICTED: similar to hCG1794790 LOC440981 0.108 PREDICTED: similar to hCG1794790 GRIN3B 0.022 glutamate receptor, ionotropic, N-methyl-D-aspartate... SEC24A 0.017 SEC24 related gene family, member A KALRN 0.013 kalirin, RhoGEF kinase isoform 1 KALRN 0.013 kalirin, RhoGEF kinase isoform 3 EP400 0.013 E1A binding protein p400 RAX 0.009 retina and anterior neural fold homeobox GORASP2 0.009 golgi reassembly stacking protein 2 MED15 0.009 mediator complex subunit 15 isoform a MED15 0.009 mediator complex subunit 15 isoform b FAM186A 0.009 family with sequence similarity 186, member A [Homo... DNAH14 0.009 dynein, axonemal, heavy polypeptide 14 isoform 1 [H... PLEKHN1 0.007 pleckstrin homology domain containing, family N mem... PLEKHN1 0.007 pleckstrin homology domain containing, family N mem... IRF5 0.007 interferon regulatory factor 5 isoform c IRF5 0.007 interferon regulatory factor 5 isoform b IRF5 0.007 interferon regulatory factor 5 isoform d IRF5 0.007 interferon regulatory factor 5 isoform c IRF5 0.007 interferon regulatory factor 5 isoform c IRF5 0.007 interferon regulatory factor 5 isoform bHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
