
| Name: HES2 | Sequence: fasta or formatted (173aa) | NCBI GI: 63055047 | |
|
Description: hairy and enhancer of split homolog 2
|
Referenced in: Notch Pathway
| ||
|
Composition:
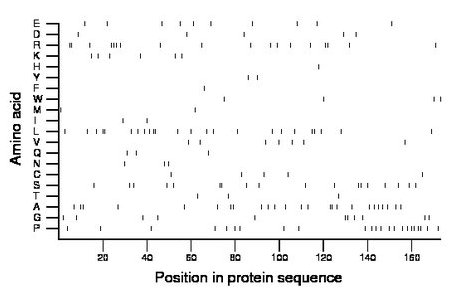
Amino acid Percentage Count Longest homopolymer A alanine 14.5 25 2 C cysteine 2.9 5 1 D aspartate 2.9 5 1 E glutamate 5.8 10 1 F phenylalanine 0.6 1 1 G glycine 6.4 11 2 H histidine 0.6 1 1 I isoleucine 1.2 2 1 K lysine 3.5 6 1 L leucine 13.9 24 2 M methionine 1.2 2 1 N asparagine 1.7 3 1 P proline 13.3 23 2 Q glutamine 1.7 3 1 R arginine 10.4 18 3 S serine 10.4 18 2 T threonine 1.7 3 1 V valine 4.0 7 1 W tryptophan 2.3 4 1 Y tyrosine 1.2 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hairy and enhancer of split homolog 2 HES4 0.290 hairy and enhancer of split 4 isoform 2 HES1 0.271 hairy and enhancer of split 1 HES4 0.265 hairy and enhancer of split 4 isoform 1 HES5 0.165 hairy and enhancer of split 5 HES6 0.159 hairy and enhancer of split 6 isoform b HES6 0.155 hairy and enhancer of split 6 isoform a HES7 0.149 hairy and enhancer of split 7 HES3 0.125 hairy and enhancer of split 3 HEY1 0.113 hairy/enhancer-of-split related with YRPW motif 1 i... HEY2 0.110 hairy/enhancer-of-split related with YRPW motif 2 [Ho... HEY1 0.107 hairy/enhancer-of-split related with YRPW motif 1 i... HEYL 0.107 hairy/enhancer-of-split related with YRPW motif-like ... BHLHE40 0.085 basic helix-loop-helix family, member e40 HCN2 0.058 hyperpolarization activated cyclic nucleotide-gated... HELT 0.055 HES/HEY-like transcription factor CDX1 0.052 caudal type homeobox 1 MAP7 0.052 microtubule-associated protein 7 MXRA7 0.052 transmembrane anchor protein 1 isoform 3 MXRA7 0.052 transmembrane anchor protein 1 isoform 2 MXRA7 0.052 transmembrane anchor protein 1 isoform 1 BHLHE41 0.049 basic helix-loop-helix domain containing, class B, 3... ZNF828 0.046 zinc finger protein 828 SEC24B 0.046 SEC24 (S. cerevisiae) homolog B isoform b SEC24B 0.046 SEC24 (S. cerevisiae) homolog B isoform a TRIM33 0.046 tripartite motif-containing 33 protein isoform beta ... TRIM33 0.046 tripartite motif-containing 33 protein isoform alpha... TRRAP 0.046 transformation/transcription domain-associated protei... LBXCOR1 0.046 LBXCOR1 homolog TRIM47 0.043 tripartite motif-containing 47Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
