
| Name: CDC73 | Sequence: fasta or formatted (531aa) | NCBI GI: 40018640 | |
|
Description: parafibromin
|
Referenced in: RNA Polymerase and General Transcription Factors
| ||
|
Composition:
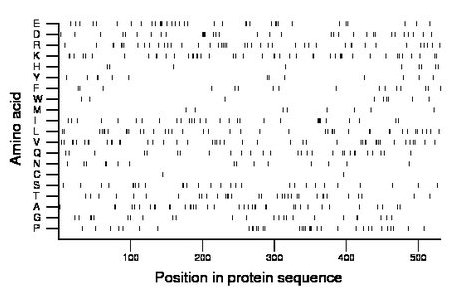
Amino acid Percentage Count Longest homopolymer A alanine 7.5 40 2 C cysteine 0.4 2 1 D aspartate 6.2 33 3 E glutamate 6.6 35 2 F phenylalanine 3.0 16 1 G glycine 4.5 24 2 H histidine 1.7 9 1 I isoleucine 5.6 30 4 K lysine 8.5 45 3 L leucine 7.7 41 2 M methionine 1.7 9 1 N asparagine 2.8 15 2 P proline 6.4 34 3 Q glutamine 5.3 28 1 R arginine 8.1 43 2 S serine 5.3 28 1 T threonine 6.2 33 2 V valine 8.3 44 2 W tryptophan 1.7 9 1 Y tyrosine 2.4 13 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 parafibromin SETD1A 0.014 SET domain containing 1A PRKCSH 0.014 protein kinase C substrate 80K-H isoform 2 PRKCSH 0.014 protein kinase C substrate 80K-H isoform 1 ARID1A 0.012 AT rich interactive domain 1A isoform b CCDC6 0.012 coiled-coil domain containing 6 CCDC88B 0.011 coiled-coil domain containing 88 CHD2 0.011 chromodomain helicase DNA binding protein 2 isoform... SRRM2 0.011 splicing coactivator subunit SRm300 SCNN1D 0.010 sodium channel, nonvoltage-gated 1, delta isoform 1... MDC1 0.010 mediator of DNA-damage checkpoint 1 MYO18A 0.010 myosin 18A isoform b MYO18A 0.010 myosin 18A isoform a SCNN1D 0.009 sodium channel, nonvoltage-gated 1, delta isoform 2 ... EVPL 0.009 envoplakin SPEN 0.008 spen homolog, transcriptional regulator MAP7D1 0.008 MAP7 domain containing 1 TFAP4 0.008 transcription factor AP-4 (activating enhancer bindin... LOC643677 0.007 hypothetical protein LOC643677 LOC643677 0.007 PREDICTED: hypothetical protein LOC643677 LOC643677 0.007 PREDICTED: hypothetical protein LOC643677 LOC643677 0.007 PREDICTED: hypothetical protein LOC643677 NOP58 0.007 NOP58 ribonucleoprotein homolog PRPF4 0.007 PRP4 pre-mRNA processing factor 4 homolog SRCAP 0.007 Snf2-related CBP activator protein KIF20B 0.007 M-phase phosphoprotein 1 TJP1 0.006 tight junction protein 1 isoform b TJP1 0.006 tight junction protein 1 isoform a ZNF438 0.006 zinc finger protein 438 isoform b ZNF438 0.006 zinc finger protein 438 isoform bHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
