
| Name: C16orf75 | Sequence: fasta or formatted (147aa) | NCBI GI: 24308245 | |
|
Description: RecQ-mediated genome instability protein 2
| Not currently referenced in the text | ||
|
Composition:
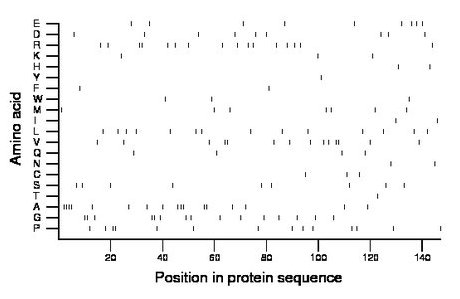
Amino acid Percentage Count Longest homopolymer A alanine 11.6 17 4 C cysteine 2.0 3 1 D aspartate 6.1 9 1 E glutamate 6.1 9 1 F phenylalanine 1.4 2 1 G glycine 10.2 15 2 H histidine 1.4 2 1 I isoleucine 1.4 2 1 K lysine 2.0 3 1 L leucine 9.5 14 1 M methionine 4.8 7 1 N asparagine 1.4 2 1 P proline 9.5 14 2 Q glutamine 2.7 4 1 R arginine 10.9 16 2 S serine 6.1 9 1 T threonine 0.7 1 1 V valine 9.5 14 2 W tryptophan 2.0 3 1 Y tyrosine 0.7 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 RecQ-mediated genome instability protein 2 LOC100292754 0.029 PREDICTED: hypothetical protein LOC100290764 0.029 PREDICTED: hypothetical protein XP_002346421 LOC100287607 0.029 PREDICTED: hypothetical protein XP_002342119 LOC100294067 0.018 PREDICTED: similar to rCG47812 MEPCE 0.018 bin3, bicoid-interacting 3 STRN4 0.018 zinedin isoform 2 STRN4 0.018 zinedin isoform 1 LOC100292196 0.014 PREDICTED: hypothetical protein C14orf80 0.014 hypothetical protein LOC283643 isoform 4 C14orf80 0.014 hypothetical protein LOC283643 isoform 2 C14orf80 0.014 hypothetical protein LOC283643 isoform 1 C2orf16 0.014 hypothetical protein LOC84226 GRIP2 0.011 glutamate receptor interacting protein 2 C2orf21 0.011 chromosome 2 open reading frame 21 isoform 1 KIAA1949 0.011 phostensin KIAA1949 0.011 phostensin AGPAT2 0.011 1-acylglycerol-3-phosphate O-acyltransferase 2 isofo... AGPAT2 0.011 1-acylglycerol-3-phosphate O-acyltransferase 2 isofor... ASPHD1 0.011 aspartate beta-hydroxylase domain containing 1 [Homo... IER5 0.007 immediate early response 5 LOC100292292 0.007 PREDICTED: hypothetical protein C18orf51 0.007 hypothetical protein LOC125704 TIE1 0.007 tyrosine kinase with immunoglobulin-like and EGF-like... ALPK3 0.007 alpha-kinase 3 KBTBD11 0.007 kelch repeat and BTB (POZ) domain containing 11 [Homo... LOC100292234 0.007 PREDICTED: hypothetical protein LOC100289952 0.007 PREDICTED: hypothetical protein XP_002348279 LOC100289949 0.007 PREDICTED: hypothetical protein XP_002346877 LOC100290708 0.007 PREDICTED: hypothetical protein XP_002346660Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
