
| Name: LOC100293573 | Sequence: fasta or formatted (77aa) | NCBI GI: 239757934 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
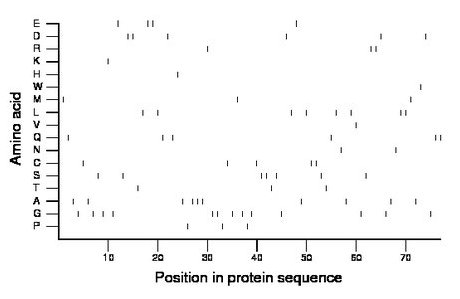
Amino acid Percentage Count Longest homopolymer A alanine 13.0 10 3 C cysteine 6.5 5 2 D aspartate 7.8 6 2 E glutamate 5.2 4 2 F phenylalanine 0.0 0 0 G glycine 16.9 13 2 H histidine 1.3 1 1 I isoleucine 0.0 0 0 K lysine 1.3 1 1 L leucine 10.4 8 2 M methionine 3.9 3 1 N asparagine 2.6 2 1 P proline 3.9 3 1 Q glutamine 7.8 6 2 R arginine 3.9 3 2 S serine 9.1 7 2 T threonine 3.9 3 1 V valine 1.3 1 1 W tryptophan 1.3 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290236 1.000 PREDICTED: hypothetical protein XP_002348273 LOC100288658 0.058 PREDICTED: hypothetical protein XP_002342750 LOC100288658 0.058 PREDICTED: hypothetical protein LOC100291624 0.036 PREDICTED: hypothetical protein XP_002345876 LOC100292753 0.036 PREDICTED: hypothetical protein LOC100289712 0.036 PREDICTED: hypothetical protein XP_002346761 LOC100290856 0.036 PREDICTED: hypothetical protein XP_002347537 LOC100287455 0.036 PREDICTED: hypothetical protein XP_002342583 LOC100289061 0.036 PREDICTED: hypothetical protein XP_002342174 LOC100131773 0.036 PREDICTED: similar to mCG116201 NHSL1 0.022 NHS-like 1 KCNH3 0.022 potassium voltage-gated channel, subfamily H (eag-re... MPND 0.022 MPN domain containing isoform 2 MPND 0.022 MPN domain containing isoform 1 LOC100293672 0.022 PREDICTED: hypothetical protein LOC100286936 0.022 PREDICTED: hypothetical protein LOC100286936 0.022 PREDICTED: hypothetical protein XP_002342699 LOR 0.022 loricrin MARCH9 0.022 membrane-associated RING-CH protein IX KAT2B 0.014 K(lysine) acetyltransferase 2B AGRN 0.014 agrin LOC100291383 0.014 PREDICTED: hypothetical protein XP_002346891 SNTB1 0.014 basic beta 1 syntrophin LOC100287812 0.014 PREDICTED: hypothetical protein XP_002343253 SMAD6 0.014 SMAD family member 6 isoform 1 GLP-1 0.014 GATA like protein-1 HRNR 0.014 hornerin LOC100128440 0.014 PREDICTED: hypothetical protein LOC100128440 0.014 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
