
| Name: LOC100293286 | Sequence: fasta or formatted (168aa) | NCBI GI: 239757060 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
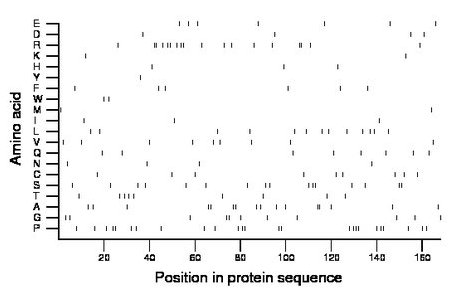
Amino acid Percentage Count Longest homopolymer A alanine 9.5 16 2 C cysteine 5.4 9 1 D aspartate 2.4 4 1 E glutamate 4.2 7 1 F phenylalanine 3.6 6 1 G glycine 6.5 11 2 H histidine 1.8 3 1 I isoleucine 1.8 3 1 K lysine 1.2 2 1 L leucine 7.7 13 1 M methionine 1.2 2 1 N asparagine 2.4 4 1 P proline 14.9 25 3 Q glutamine 4.8 8 1 R arginine 10.7 18 2 S serine 9.5 16 2 T threonine 5.4 9 1 V valine 5.4 9 1 W tryptophan 1.2 2 1 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290832 0.997 PREDICTED: hypothetical protein XP_002347902 LOC100289128 0.997 PREDICTED: hypothetical protein XP_002343651 WIPF2 0.030 WIRE protein TNKS1BP1 0.024 tankyrase 1-binding protein 1 MAST2 0.024 microtubule associated serine/threonine kinase 2 [H... PCNT 0.021 pericentrin LOC731282 0.021 PREDICTED: hypothetical protein LOC100134152 0.018 PREDICTED: similar to synaptotagmin XV-b LOC285141 0.018 PREDICTED: hypothetical protein LOC285141 LOC285141 0.018 PREDICTED: hypothetical protein LOC285141 LOC285141 0.018 PREDICTED: hypothetical protein LOC285141 MIA3 0.018 melanoma inhibitory activity family, member 3 [Homo... FAM47A 0.018 hypothetical protein LOC158724 SYT15 0.018 synaptotagmin XV isoform a SYT15 0.018 synaptotagmin XV isoform b LOC100288303 0.018 PREDICTED: hypothetical protein XP_002343794 LOC731282 0.015 PREDICTED: hypothetical protein INF2 0.015 inverted formin 2 isoform 2 INF2 0.015 inverted formin 2 isoform 1 TMPRSS13 0.015 transmembrane protease, serine 13 KALRN 0.015 kalirin, RhoGEF kinase isoform 1 KALRN 0.015 kalirin, RhoGEF kinase isoform 2 LOC100288243 0.015 PREDICTED: hypothetical protein LOC100288243 0.015 PREDICTED: hypothetical protein XP_002342233 IGHMBP2 0.012 immunoglobulin mu binding protein 2 LOC100293644 0.012 PREDICTED: hypothetical protein LOC100287563 0.012 PREDICTED: hypothetical protein LOC100287563 0.012 PREDICTED: hypothetical protein XP_002343733 GLTSCR1 0.012 glioma tumor suppressor candidate region gene 1 [Ho...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
