
| Name: LOC100293652 | Sequence: fasta or formatted (281aa) | NCBI GI: 239756833 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
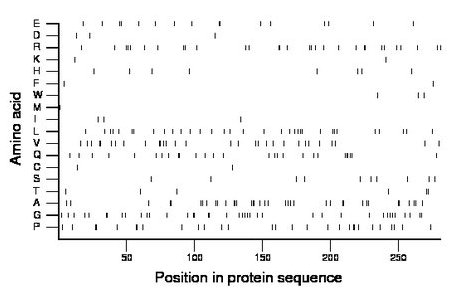
Amino acid Percentage Count Longest homopolymer A alanine 13.2 37 3 C cysteine 0.7 2 1 D aspartate 1.1 3 1 E glutamate 5.7 16 2 F phenylalanine 0.7 2 1 G glycine 13.9 39 2 H histidine 2.8 8 1 I isoleucine 1.1 3 1 K lysine 0.7 2 1 L leucine 11.7 33 2 M methionine 0.4 1 1 N asparagine 0.0 0 0 P proline 10.3 29 2 Q glutamine 9.6 27 3 R arginine 10.7 30 2 S serine 3.6 10 1 T threonine 2.1 6 2 V valine 10.7 30 2 W tryptophan 1.1 3 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100291016 0.993 PREDICTED: hypothetical protein XP_002347804 LOC100287809 0.993 PREDICTED: hypothetical protein XP_002343597 LOC100132168 0.101 PREDICTED: hypothetical protein LOC100292754 0.041 PREDICTED: hypothetical protein LOC100290764 0.041 PREDICTED: hypothetical protein XP_002346421 LOC100287607 0.041 PREDICTED: hypothetical protein XP_002342119 LOC728767 0.026 PREDICTED: hypothetical protein LOC728767 0.026 PREDICTED: hypothetical protein CACNA1B 0.024 calcium channel, voltage-dependent, N type, alpha 1B ... GLTSCR1 0.024 glioma tumor suppressor candidate region gene 1 [Ho... LOC100292595 0.022 PREDICTED: hypothetical protein LOC100290895 0.022 PREDICTED: hypothetical protein XP_002348072 LOC100287206 0.022 PREDICTED: hypothetical protein XP_002343750 LOC100293492 0.022 PREDICTED: hypothetical protein LOC729162 0.021 PREDICTED: similar to hCG1981372 LOC729162 0.021 PREDICTED: similar to hCG1981372 LOC100288030 0.021 PREDICTED: hypothetical protein XP_002343812 LOC375295 0.021 PREDICTED: hypothetical protein LOC389906 0.019 PREDICTED: similar to Serine/threonine-protein kina... APC2 0.019 adenomatosis polyposis coli 2 HES1 0.019 hairy and enhancer of split 1 LOC100288108 0.019 PREDICTED: hypothetical protein LOC100288108 0.019 PREDICTED: hypothetical protein XP_002342919 GPR162 0.019 G protein-coupled receptor 162 isoform 2 GPR162 0.019 G protein-coupled receptor 162 isoform 1 LOC100293818 0.019 PREDICTED: hypothetical protein LOC100132799 0.019 PREDICTED: hypothetical protein DDN 0.019 dendrin SUPT6H 0.019 suppressor of Ty 6 homologHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
