
| Name: LOC100292168 | Sequence: fasta or formatted (282aa) | NCBI GI: 239754378 | |
|
Description: PREDICTED: similar to hCG2030094
| Not currently referenced in the text | ||
|
Composition:
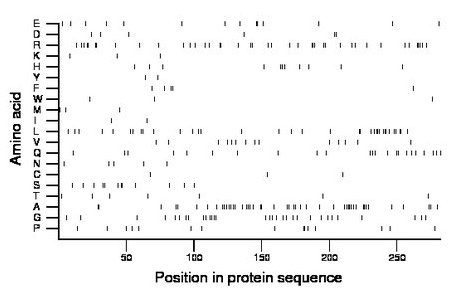
Amino acid Percentage Count Longest homopolymer A alanine 16.7 47 4 C cysteine 1.1 3 1 D aspartate 2.1 6 2 E glutamate 5.0 14 2 F phenylalanine 1.8 5 2 G glycine 11.0 31 2 H histidine 3.9 11 2 I isoleucine 0.7 2 1 K lysine 1.1 3 1 L leucine 12.4 35 3 M methionine 1.1 3 1 N asparagine 1.8 5 1 P proline 5.3 15 2 Q glutamine 8.2 23 2 R arginine 14.5 41 3 S serine 4.3 12 2 T threonine 2.5 7 1 V valine 5.0 14 1 W tryptophan 1.1 3 1 Y tyrosine 0.7 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: similar to hCG2030094 LOC100288743 0.864 PREDICTED: similar to hCG2030094 LOC100288743 0.864 PREDICTED: hypothetical protein XP_002342716 LOC100289993 0.041 PREDICTED: hypothetical protein XP_002348146 LOC100288915 0.041 PREDICTED: hypothetical protein XP_002343820 LOC728767 0.037 PREDICTED: hypothetical protein LOC728767 0.037 PREDICTED: hypothetical protein LOC729154 0.026 PREDICTED: hypothetical protein LOC729154 0.026 PREDICTED: hypothetical protein HOXD11 0.026 homeobox D11 LOC100290372 0.022 PREDICTED: hypothetical protein XP_002346788 LOC100288606 0.022 PREDICTED: hypothetical protein XP_002342652 LOC100132983 0.022 PREDICTED: hypothetical protein LOC100288686 0.019 PREDICTED: hypothetical protein XP_002342615 LOC100294413 0.019 PREDICTED: hypothetical protein APC2 0.019 adenomatosis polyposis coli 2 SOX12 0.019 SRY (sex determining region Y)-box 12 CNGB1 0.017 cyclic nucleotide gated channel beta 1 isoform a [H... NOM1 0.017 nucleolar protein with MIF4G domain 1 LOC100127944 0.015 PREDICTED: hypothetical protein LOC100127944 0.015 PREDICTED: hypothetical protein LOC100127944 0.015 PREDICTED: hypothetical protein BRSK1 0.015 BR serine/threonine kinase 1 KIAA0913 0.015 hypothetical protein LOC23053 LOC100292017 0.015 PREDICTED: hypothetical protein XP_002344620 LOC100292292 0.015 PREDICTED: hypothetical protein LOC100289978 0.015 PREDICTED: hypothetical protein XP_002348035 LOC100291384 0.015 PREDICTED: hypothetical protein XP_002347156 LOC100289716 0.015 PREDICTED: hypothetical protein XP_002346898 LOC100287578 0.015 PREDICTED: hypothetical protein XP_002343004Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
