
| Name: LOC100292017 | Sequence: fasta or formatted (102aa) | NCBI GI: 239755607 | |
|
Description: PREDICTED: hypothetical protein XP_002344620
| Not currently referenced in the text | ||
|
Composition:
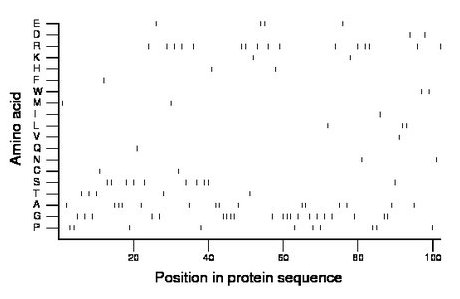
Amino acid Percentage Count Longest homopolymer A alanine 14.7 15 3 C cysteine 2.0 2 1 D aspartate 2.0 2 1 E glutamate 3.9 4 2 F phenylalanine 1.0 1 1 G glycine 20.6 21 4 H histidine 2.0 2 1 I isoleucine 1.0 1 1 K lysine 2.0 2 1 L leucine 2.9 3 2 M methionine 2.0 2 1 N asparagine 2.0 2 1 P proline 9.8 10 2 Q glutamine 1.0 1 1 R arginine 15.7 16 2 S serine 9.8 10 2 T threonine 4.9 5 1 V valine 1.0 1 1 W tryptophan 2.0 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002344620 LOC100291634 0.094 PREDICTED: hypothetical protein XP_002346062 LOC100291096 0.094 PREDICTED: hypothetical protein XP_002346952 LOC100287232 0.094 PREDICTED: hypothetical protein XP_002342816 LOC100287232 0.094 PREDICTED: hypothetical protein LOC100291076 0.094 PREDICTED: hypothetical protein XP_002347846 MBD2 0.089 methyl-CpG binding domain protein 2 testis-specific i... MBD2 0.089 methyl-CpG binding domain protein 2 isoform 1 FLJ10357 0.073 hypothetical protein LOC55701 LOC100293375 0.068 PREDICTED: hypothetical protein NOTCH4 0.062 notch4 preproprotein LOC100293922 0.057 PREDICTED: hypothetical protein LOC100292773 0.057 PREDICTED: hypothetical protein LOC100291358 0.057 PREDICTED: hypothetical protein XP_002347390 LOC100290063 0.057 PREDICTED: hypothetical protein XP_002346992 LOC100287283 0.057 PREDICTED: hypothetical protein XP_002342840 LOC100287283 0.057 PREDICTED: hypothetical protein FOXD2 0.057 forkhead box D2 LOC100292082 0.057 PREDICTED: hypothetical protein XP_002345881 GLCCI1 0.057 glucocorticoid induced transcript 1 OSBP 0.057 oxysterol binding protein SORCS2 0.052 VPS10 domain receptor protein SORCS 2 HRNR 0.052 hornerin LOC100293975 0.052 PREDICTED: hypothetical protein LOC100290812 0.052 PREDICTED: hypothetical protein XP_002347678 LOC100288007 0.052 PREDICTED: hypothetical protein LOC100288205 0.052 PREDICTED: hypothetical protein XP_002343496 LOC100288382 0.052 PREDICTED: hypothetical protein XP_002342213 LOC100288007 0.052 PREDICTED: hypothetical protein XP_002342192 PAK4 0.052 p21-activated kinase 4 isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
