
| Name: LOC100293980 | Sequence: fasta or formatted (213aa) | NCBI GI: 239753500 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
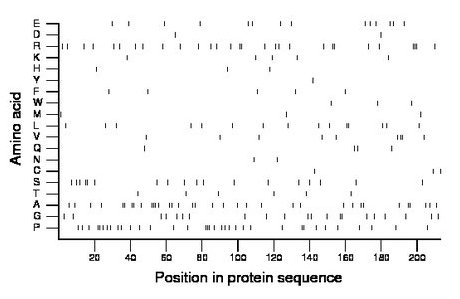
Amino acid Percentage Count Longest homopolymer A alanine 16.4 35 3 C cysteine 1.4 3 1 D aspartate 0.9 2 1 E glutamate 6.6 14 1 F phenylalanine 2.3 5 1 G glycine 9.9 21 2 H histidine 1.4 3 1 I isoleucine 0.0 0 0 K lysine 2.3 5 1 L leucine 7.0 15 2 M methionine 1.4 3 1 N asparagine 0.9 2 1 P proline 16.4 35 3 Q glutamine 1.9 4 1 R arginine 13.6 29 3 S serine 8.5 18 2 T threonine 2.8 6 1 V valine 4.2 9 2 W tryptophan 1.4 3 1 Y tyrosine 0.5 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100290741 0.995 PREDICTED: hypothetical protein XP_002346521 LOC100288445 0.995 PREDICTED: hypothetical protein XP_002342359 SAMD1 0.075 sterile alpha motif domain containing 1 FAM71E2 0.053 hypothetical protein LOC284418 RNF19B 0.053 ring finger protein 19B isoform a RNF19B 0.053 ring finger protein 19B isoform b NACA 0.053 nascent polypeptide-associated complex alpha subuni... ARAP1 0.053 ArfGAP with RhoGAP domain, ankyrin repeat and PH dom... KAT2A 0.053 general control of amino acid synthesis 5-like 2 [H... PTPN23 0.053 protein tyrosine phosphatase, non-receptor type 23 [... LOC645321 0.051 PREDICTED: hypothetical protein LOC645321 0.051 PREDICTED: hypothetical protein TAF4 0.051 TBP-associated factor 4 SRRM1 0.051 serine/arginine repetitive matrix 1 LOC100130360 0.048 PREDICTED: hypothetical protein MAMSTR 0.048 MEF2 activating motif and SAP domain containing tra... MAMSTR 0.048 MEF2 activating motif and SAP domain containing tran... ZBTB4 0.048 zinc finger and BTB domain containing 4 ZBTB4 0.048 zinc finger and BTB domain containing 4 RICH2 0.048 Rho GTPase-activating protein RICH2 SYNPO2L 0.048 synaptopodin 2-like isoform a SYNPO2L 0.048 synaptopodin 2-like isoform b LOC100131102 0.048 PREDICTED: hypothetical protein LOC100131102 0.048 PREDICTED: hypothetical protein LOC729370 0.048 PREDICTED: hypothetical protein ZNF828 0.046 zinc finger protein 828 CDKN1C 0.046 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.046 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.046 cyclin-dependent kinase inhibitor 1C isoform aHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
