
| Name: LOC100293546 | Sequence: fasta or formatted (141aa) | NCBI GI: 239753002 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
|
Composition:
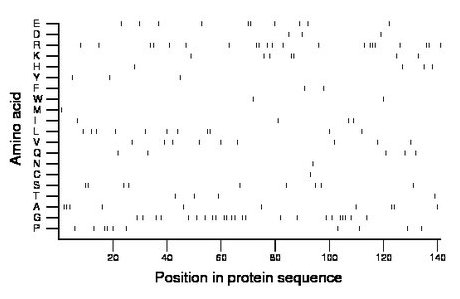
Amino acid Percentage Count Longest homopolymer A alanine 7.1 10 3 C cysteine 0.7 1 1 D aspartate 2.1 3 1 E glutamate 7.1 10 2 F phenylalanine 1.4 2 1 G glycine 17.0 24 3 H histidine 2.8 4 1 I isoleucine 2.8 4 1 K lysine 5.0 7 2 L leucine 7.8 11 2 M methionine 0.7 1 1 N asparagine 0.7 1 1 P proline 7.1 10 2 Q glutamine 3.5 5 1 R arginine 14.9 21 3 S serine 6.4 9 2 T threonine 2.8 4 1 V valine 6.4 9 1 W tryptophan 1.4 2 1 Y tyrosine 2.1 3 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC100292407 0.026 PREDICTED: hypothetical protein LOC100293672 0.023 PREDICTED: hypothetical protein LOC100286936 0.023 PREDICTED: hypothetical protein LOC100286936 0.023 PREDICTED: hypothetical protein XP_002342699 DSG1 0.023 desmoglein 1 preproprotein HRNR 0.023 hornerin LOC100129463 0.019 PREDICTED: hypothetical protein LOC100129463 0.019 PREDICTED: hypothetical protein LOC100129463 0.019 PREDICTED: hypothetical protein LOC100129463 0.019 PREDICTED: hypothetical protein LOC649201 0.015 PREDICTED: similar to paraneoplastic antigen MA1 [H... TAF15 0.015 TBP-associated factor 15 isoform 2 TAF15 0.015 TBP-associated factor 15 isoform 1 C6orf174 0.015 hypothetical protein LOC387104 ZNF716 0.015 PREDICTED: zinc finger protein 716 LOC100287812 0.011 PREDICTED: hypothetical protein XP_002343253 C1QTNF7 0.011 C1q and tumor necrosis factor related protein 7 isof... C1QTNF7 0.011 C1q and tumor necrosis factor related protein 7 iso... C1QTNF7 0.011 C1q and tumor necrosis factor related protein 7 iso... KRT10 0.011 keratin 10 KIAA0802 0.011 hypothetical protein LOC23255 LOC728767 0.011 PREDICTED: hypothetical protein LOC728767 0.011 PREDICTED: hypothetical protein COL8A2 0.011 collagen, type VIII, alpha 2 TIMELESS 0.011 timeless homolog AHNAK2 0.011 AHNAK nucleoprotein 2 CACNG8 0.011 voltage-dependent calcium channel gamma-8 subunit [H... LOC649201 0.008 PREDICTED: similar to paraneoplastic antigen MA1 [H... UBAP2L 0.008 ubiquitin associated protein 2-like isoform a [Homo...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
