
| Name: LOC100291447 | Sequence: fasta or formatted (220aa) | NCBI GI: 239752263 | |
|
Description: PREDICTED: hypothetical protein XP_002348185
| Not currently referenced in the text | ||
|
Composition:
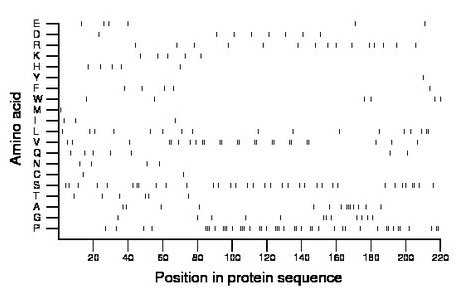
Amino acid Percentage Count Longest homopolymer A alanine 6.4 14 3 C cysteine 0.9 2 1 D aspartate 3.6 8 1 E glutamate 2.7 6 1 F phenylalanine 2.3 5 1 G glycine 5.5 12 2 H histidine 2.3 5 1 I isoleucine 1.4 3 1 K lysine 2.3 5 1 L leucine 7.7 17 2 M methionine 0.5 1 1 N asparagine 1.8 4 1 P proline 19.5 43 3 Q glutamine 3.2 7 1 R arginine 6.8 15 1 S serine 15.9 35 2 T threonine 2.7 6 1 V valine 11.4 25 2 W tryptophan 2.7 6 1 Y tyrosine 0.5 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002348185 LOC100288699 0.620 PREDICTED: hypothetical protein XP_002343846 LOC100291479 0.604 PREDICTED: hypothetical protein XP_002348186 LOC100293936 0.316 PREDICTED: hypothetical protein LOC100287915 0.316 PREDICTED: hypothetical protein XP_002343907 LOC100287882 0.316 PREDICTED: hypothetical protein XP_002343906 PCLO 0.094 piccolo isoform 2 PCLO 0.094 piccolo isoform 1 YLPM1 0.090 YLP motif containing 1 SYVN1 0.078 synoviolin 1 isoform a FLJ22184 0.078 PREDICTED: hypothetical protein FLJ22184 TAF4 0.076 TBP-associated factor 4 SYVN1 0.076 synoviolin 1 isoform b SF1 0.071 splicing factor 1 isoform 1 KIAA1522 0.071 hypothetical protein LOC57648 PRB1 0.069 proline-rich protein BstNI subfamily 1 isoform 1 pre... SF1 0.069 splicing factor 1 isoform 2 PRB2 0.069 proline-rich protein BstNI subfamily 2 DIAPH1 0.067 diaphanous 1 isoform 2 DIAPH1 0.067 diaphanous 1 isoform 1 FLJ22184 0.065 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.065 PREDICTED: hypothetical protein LOC80164 SAMD1 0.065 sterile alpha motif domain containing 1 FMN1 0.062 formin 1 SOS1 0.062 son of sevenless homolog 1 PRB1 0.062 proline-rich protein BstNI subfamily 1 isoform 2 pre... HCN2 0.062 hyperpolarization activated cyclic nucleotide-gated... SFPQ 0.062 splicing factor proline/glutamine rich (polypyrimidin... WASL 0.060 Wiskott-Aldrich syndrome gene-like protein APBB1IP 0.060 amyloid beta (A4) precursor protein-binding, family ...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
