
| Name: LOC100290029 | Sequence: fasta or formatted (132aa) | NCBI GI: 239751067 | |
|
Description: PREDICTED: hypothetical protein XP_002347693
| Not currently referenced in the text | ||
|
Composition:
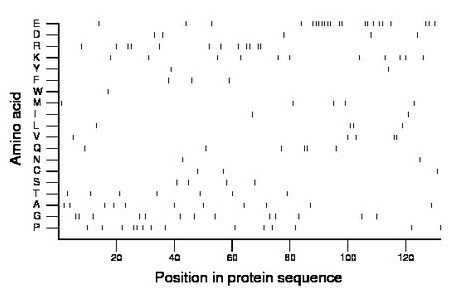
Amino acid Percentage Count Longest homopolymer A alanine 8.3 11 1 C cysteine 2.3 3 1 D aspartate 3.8 5 1 E glutamate 16.7 22 7 F phenylalanine 2.3 3 1 G glycine 9.8 13 2 H histidine 0.0 0 0 I isoleucine 1.5 2 1 K lysine 8.3 11 1 L leucine 3.0 4 2 M methionine 3.8 5 1 N asparagine 1.5 2 1 P proline 10.6 14 2 Q glutamine 4.5 6 2 R arginine 9.1 12 2 S serine 3.0 4 1 T threonine 5.3 7 1 V valine 3.8 5 2 W tryptophan 0.8 1 1 Y tyrosine 1.5 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002347693 ZNF771 0.287 zinc finger protein 771 ZNF771 0.287 zinc finger protein 771 C3orf44 0.088 hypothetical protein LOC131831 LOC731605 0.060 PREDICTED: similar to BCL2-associated transcription... KLHDC7B 0.060 kelch domain containing 7B NEFH 0.056 neurofilament, heavy polypeptide 200kDa SCAF1 0.056 SR-related CTD-associated factor 1 PRB3 0.056 proline-rich protein BstNI subfamily 3 precursor [H... PPARGC1B 0.052 peroxisome proliferator-activated receptor gamma, co... TULP1 0.052 tubby like protein 1 ATN1 0.048 atrophin-1 ATN1 0.048 atrophin-1 ZFHX3 0.048 AT-binding transcription factor 1 STK11IP 0.048 LKB1 interacting protein LOC731605 0.044 PREDICTED: similar to BCL2-associated transcription... BCLAF1 0.044 BCL2-associated transcription factor 1 isoform 3 [H... BCLAF1 0.044 BCL2-associated transcription factor 1 isoform 2 [H... BCLAF1 0.044 BCL2-associated transcription factor 1 isoform 1 [Hom... KIAA1429 0.044 hypothetical protein LOC25962 isoform 1 KIAA1429 0.044 hypothetical protein LOC25962 isoform 2 PHF23 0.040 PHD finger protein 23 TTBK1 0.040 tau tubulin kinase 1 SYN2 0.040 synapsin II isoform IIa SRRM2 0.040 splicing coactivator subunit SRm300 BAZ1B 0.040 bromodomain adjacent to zinc finger domain, 1B [Homo... LOC100289089 0.040 PREDICTED: hypothetical protein XP_002343359 MLL4 0.040 myeloid/lymphoid or mixed-lineage leukemia 4 LOC100133758 0.036 PREDICTED: hypothetical protein, partial BSN 0.036 bassoon proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
