
| Name: LOC100290693 | Sequence: fasta or formatted (178aa) | NCBI GI: 239749157 | |
|
Description: PREDICTED: hypothetical protein XP_002346930
| Not currently referenced in the text | ||
|
Composition:
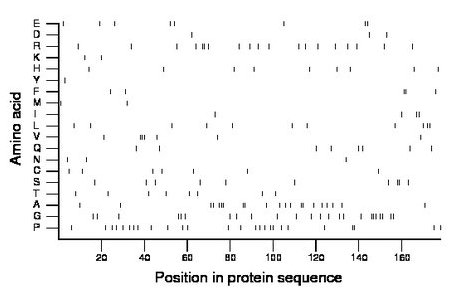
Amino acid Percentage Count Longest homopolymer A alanine 11.8 21 3 C cysteine 3.9 7 1 D aspartate 1.7 3 1 E glutamate 4.5 8 2 F phenylalanine 2.8 5 2 G glycine 13.5 24 3 H histidine 5.1 9 1 I isoleucine 2.2 4 2 K lysine 1.1 2 1 L leucine 6.2 11 2 M methionine 1.1 2 1 N asparagine 1.7 3 1 P proline 14.6 26 2 Q glutamine 4.5 8 1 R arginine 10.7 19 2 S serine 5.6 10 2 T threonine 4.5 8 1 V valine 3.9 7 3 W tryptophan 0.0 0 0 Y tyrosine 0.6 1 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002346930 LOC100289070 1.000 PREDICTED: hypothetical protein XP_002342806 LOC100289070 1.000 PREDICTED: hypothetical protein LOC100293115 0.986 PREDICTED: hypothetical protein ZFR2 0.050 zinc finger RNA binding protein 2 isoform 1 LOC284988 0.048 PREDICTED: hypothetical protein NKX1-1 0.048 PREDICTED: HPX-153 homeobox HCN2 0.045 hyperpolarization activated cyclic nucleotide-gated... LOC100290612 0.045 PREDICTED: hypothetical protein XP_002348127 BBC3 0.042 BCL2 binding component 3 isoform 1 LOC100289242 0.042 PREDICTED: hypothetical protein XP_002342243 SEMA6C 0.042 semaphorin Y C5orf33 0.042 hypothetical protein LOC133686 isoform 1 SFPQ 0.042 splicing factor proline/glutamine rich (polypyrimidin... NKX1-1 0.039 PREDICTED: NK1 homeobox 1 GNAS 0.039 GNAS complex locus XLas GGN 0.039 gametogenetin CACNA1A 0.039 calcium channel, alpha 1A subunit isoform 4 VASP 0.036 vasodilator-stimulated phosphoprotein C1orf229 0.036 hypothetical protein LOC388759 TBKBP1 0.036 TBK1 binding protein 1 LOC100291243 0.036 PREDICTED: hypothetical protein XP_002348077 LOC100134105 0.034 PREDICTED: similar to hCG1793472 SF3B4 0.034 splicing factor 3b, subunit 4 SCARF2 0.034 scavenger receptor class F, member 2 isoform 1 [Homo... SCARF2 0.034 scavenger receptor class F, member 2 isoform 2 [Homo... RBM15 0.034 RNA binding motif protein 15 SAMD1 0.034 sterile alpha motif domain containing 1 LOC100293129 0.034 PREDICTED: hypothetical protein BEND4 0.034 BEN domain containing 4 isoform bHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
