
| Name: LOC100286931 | Sequence: fasta or formatted (72aa) | NCBI GI: 239743889 | |
|
Description: PREDICTED: hypothetical protein XP_002342998
| Not currently referenced in the text | ||
|
Composition:
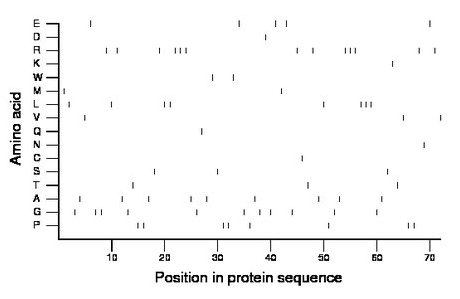
Amino acid Percentage Count Longest homopolymer A alanine 12.5 9 1 C cysteine 1.4 1 1 D aspartate 1.4 1 1 E glutamate 6.9 5 1 F phenylalanine 0.0 0 0 G glycine 15.3 11 2 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 1.4 1 1 L leucine 11.1 8 3 M methionine 2.8 2 1 N asparagine 1.4 1 1 P proline 11.1 8 2 Q glutamine 1.4 1 1 R arginine 18.1 13 3 S serine 4.2 3 1 T threonine 4.2 3 1 V valine 4.2 3 1 W tryptophan 2.8 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342998 LOC100291856 0.952 PREDICTED: hypothetical protein XP_002346216 LOC100288067 0.952 PREDICTED: hypothetical protein XP_002342969 LOC100291961 0.063 PREDICTED: hypothetical protein XP_002345135 LOC100291996 0.056 PREDICTED: hypothetical protein XP_002345433 LOC100289507 0.056 PREDICTED: hypothetical protein LOC100289507 0.056 PREDICTED: hypothetical protein XP_002344219 LOC100129781 0.048 PREDICTED: hypothetical protein PELP1 0.048 proline, glutamic acid and leucine rich protein 1 [... LOC100127930 0.032 PREDICTED: hypothetical protein LOC100127930 0.032 PREDICTED: hypothetical protein ZNF133 0.032 zinc finger protein 133 ZNF133 0.032 zinc finger protein 133 MDN1 0.032 MDN1, midasin homolog LOC100134152 0.024 PREDICTED: similar to synaptotagmin XV-b JMJD8 0.024 jumonji domain containing 8 SPEG 0.016 SPEG complex locus ARMC5 0.016 armadillo repeat containing 5 isoform b ARMC5 0.016 armadillo repeat containing 5 isoform a LOC100129697 0.016 PREDICTED: hypothetical protein LOC643355 0.016 PREDICTED: hypothetical protein LOC100291147 0.016 PREDICTED: hypothetical protein XP_002347883 LOC643355 0.016 PREDICTED: hypothetical protein LOC100289560 0.016 PREDICTED: hypothetical protein XP_002344188 LOC643355 0.016 PREDICTED: hypothetical protein TFR2 0.016 transferrin receptor 2 DYNC2H1 0.016 dynein, cytoplasmic 2, heavy chain 1 LOC729575 0.016 PREDICTED: hypothetical protein LOC100288110 0.016 PREDICTED: hypothetical protein LOC100129697 0.016 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
