
| Name: JMJD8 | Sequence: fasta or formatted (285aa) | NCBI GI: 56090146 | |
|
Description: jumonji domain containing 8
|
Referenced in: Histones, Related Proteins, and Modifying Enzymes
| ||
|
Composition:
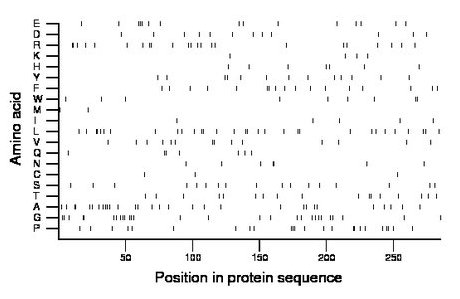
Amino acid Percentage Count Longest homopolymer A alanine 10.5 30 2 C cysteine 1.1 3 1 D aspartate 4.2 12 1 E glutamate 5.3 15 3 F phenylalanine 6.0 17 2 G glycine 9.8 28 2 H histidine 2.5 7 1 I isoleucine 1.8 5 1 K lysine 1.4 4 1 L leucine 10.9 31 2 M methionine 0.7 2 1 N asparagine 2.5 7 2 P proline 8.4 24 3 Q glutamine 2.5 7 2 R arginine 8.1 23 2 S serine 6.0 17 1 T threonine 6.0 17 2 V valine 4.9 14 2 W tryptophan 3.2 9 2 Y tyrosine 4.6 13 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 jumonji domain containing 8 JMJD6 0.069 jumonji domain containing 6 isoform 1 JMJD6 0.069 jumonji domain containing 6 isoform 2 JMJD4 0.052 jumonji domain containing 4 isoform 2 JMJD4 0.052 jumonji domain containing 4 isoform 1 HSPBAP1 0.047 Hspb associated protein 1 JMJD5 0.033 jumonji domain containing 5 isoform 1 JMJD5 0.033 jumonji domain containing 5 isoform 2 C2orf60 0.021 hypothetical protein LOC129450 JMJD7 0.019 jumonji domain containing 7 HIF1AN 0.014 hypoxia-inducible factor 1, alpha subunit inhibitor... ST5 0.012 suppression of tumorigenicity 5 isoform 1 ST5 0.012 suppression of tumorigenicity 5 isoform 1 LOC730036 0.012 PREDICTED: hypothetical protein MYO15A 0.010 myosin XV BPTF 0.010 bromodomain PHD finger transcription factor isoform ... BPTF 0.010 bromodomain PHD finger transcription factor isoform ... LOC100288928 0.010 PREDICTED: hypothetical protein LOC100288928 0.010 PREDICTED: hypothetical protein XP_002342353 LAMB4 0.010 laminin, beta 4 LOC100287450 0.009 PREDICTED: hypothetical protein XP_002343550 LOC100287417 0.009 PREDICTED: hypothetical protein XP_002343549 LOC100287374 0.009 PREDICTED: hypothetical protein XP_002343548 SMR3A 0.009 submaxillary gland androgen regulated protein 3 homo... SMR3B 0.009 submaxillary gland androgen regulated protein 3 homol... HOXA13 0.007 homeobox A13 FAM22A 0.007 hypothetical protein LOC728118 LOC100292088 0.007 PREDICTED: hypothetical protein XP_002346321 FAM22E 0.007 PREDICTED: hypothetical protein FAM22B 0.007 PREDICTED: hypothetical protein isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
