
| Name: LOC100288697 | Sequence: fasta or formatted (67aa) | NCBI GI: 239742332 | |
|
Description: PREDICTED: hypothetical protein XP_002342494
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {67aa} PREDICTED: hypothetical protein | |||
|
Composition:
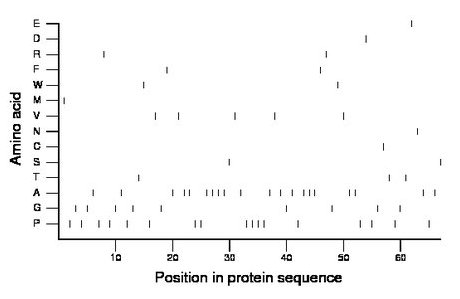
Amino acid Percentage Count Longest homopolymer A alanine 29.9 20 4 C cysteine 1.5 1 1 D aspartate 1.5 1 1 E glutamate 1.5 1 1 F phenylalanine 3.0 2 1 G glycine 13.4 9 1 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 0.0 0 0 L leucine 0.0 0 0 M methionine 1.5 1 1 N asparagine 1.5 1 1 P proline 25.4 17 4 Q glutamine 0.0 0 0 R arginine 3.0 2 1 S serine 3.0 2 1 T threonine 4.5 3 1 V valine 7.5 5 1 W tryptophan 3.0 2 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342494 LOC100293375 1.000 PREDICTED: hypothetical protein LOC100288697 1.000 PREDICTED: hypothetical protein CDKN1C 0.139 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.139 cyclin-dependent kinase inhibitor 1C isoform b [Hom... CDKN1C 0.139 cyclin-dependent kinase inhibitor 1C isoform a SAMD1 0.139 sterile alpha motif domain containing 1 PHOX2B 0.131 paired-like homeobox 2b TAF4 0.107 TBP-associated factor 4 DMRT3 0.107 doublesex and mab-3 related transcription factor 3 [... FOXD2 0.107 forkhead box D2 LGALS3 0.098 galectin 3 KIF13B 0.098 kinesin family member 13B COL15A1 0.098 alpha 1 type XV collagen precursor STK39 0.098 serine threonine kinase 39 (STE20/SPS1 homolog, yea... FAM48B1 0.098 hypothetical protein LOC100130302 DIAPH1 0.090 diaphanous 1 isoform 2 DIAPH1 0.090 diaphanous 1 isoform 1 EIF3F 0.090 eukaryotic translation initiation factor 3, subunit 5... COL1A1 0.082 alpha 1 type I collagen preproprotein LOC100128074 0.082 PREDICTED: hypothetical protein BCORL1 0.082 BCL6 co-repressor-like 1 ECOP 0.082 EGFR-coamplified and overexpressed protein SYNJ1 0.082 synaptojanin 1 isoform a SYNJ1 0.082 synaptojanin 1 isoform b LOC729086 0.082 PREDICTED: similar to EGFR-coamplified and overexpr... LOC729086 0.082 PREDICTED: similar to EGFR-coamplified and overexpr... FMNL1 0.082 formin-like 1 C9orf86 0.082 Rab-like GTP-binding protein 1 isoform 1 PLSCR1 0.082 phospholipid scramblase 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
