
| Name: LOC284988 | Sequence: fasta or formatted (53aa) | NCBI GI: 239741762 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {113aa} PREDICTED: hypothetical protein | |||
|
Composition:
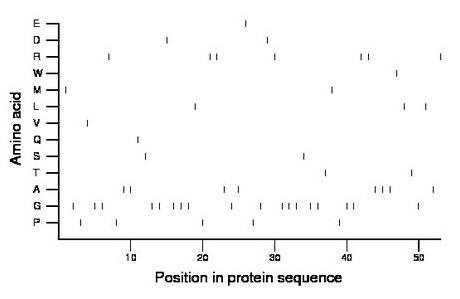
Amino acid Percentage Count Longest homopolymer A alanine 15.1 8 3 C cysteine 0.0 0 0 D aspartate 3.8 2 1 E glutamate 1.9 1 1 F phenylalanine 0.0 0 0 G glycine 34.0 18 3 H histidine 0.0 0 0 I isoleucine 0.0 0 0 K lysine 0.0 0 0 L leucine 5.7 3 1 M methionine 3.8 2 1 N asparagine 0.0 0 0 P proline 9.4 5 1 Q glutamine 1.9 1 1 R arginine 13.2 7 2 S serine 3.8 2 1 T threonine 3.8 2 1 V valine 1.9 1 1 W tryptophan 1.9 1 1 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC284988 0.856 PREDICTED: hypothetical protein POU4F1 0.144 POU domain, class 4, transcription factor 1 ZNF282 0.133 zinc finger protein 282 TAF15 0.111 TBP-associated factor 15 isoform 2 TAF15 0.111 TBP-associated factor 15 isoform 1 LOC100293176 0.111 PREDICTED: hypothetical protein LOC100289054 0.111 PREDICTED: hypothetical protein LOC100289054 0.111 PREDICTED: hypothetical protein XP_002343716 FZD8 0.100 frizzled 8 CASC3 0.089 metastatic lymph node 51 YEATS2 0.089 YEATS domain containing 2 KHSRP 0.089 KH-type splicing regulatory protein DHX36 0.089 DEAH (Asp-Glu-Ala-His) box polypeptide 36 isoform 2... DHX36 0.089 DEAH (Asp-Glu-Ala-His) box polypeptide 36 isoform 1... KRT10 0.089 keratin 10 GAR1 0.089 nucleolar protein family A, member 1 GAR1 0.089 nucleolar protein family A, member 1 C2orf21 0.089 chromosome 2 open reading frame 21 isoform 1 KRT9 0.089 keratin 9 SLC4A5 0.089 sodium bicarbonate transporter 4 isoform a SLC4A5 0.089 sodium bicarbonate transporter 4 isoform c MAF 0.089 v-maf musculoaponeurotic fibrosarcoma oncogene homol... MAF 0.089 v-maf musculoaponeurotic fibrosarcoma oncogene homolo... GSK3A 0.089 glycogen synthase kinase 3 alpha NRXN1 0.078 neurexin 1 isoform beta precursor FAM98A 0.078 hypothetical protein LOC25940 SHOX2 0.078 short stature homeobox 2 isoform a PCDH7 0.078 protocadherin 7 isoform b precursor PCDH7 0.078 protocadherin 7 isoform c precursorHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
