
| Name: LOC100288570 | Sequence: fasta or formatted (248aa) | NCBI GI: 239741718 | |
|
Description: PREDICTED: hypothetical protein XP_002342296
|
Referenced in:
| ||
|
Composition:
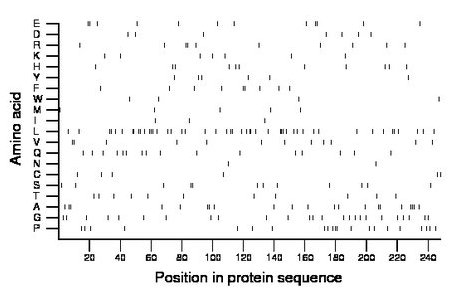
Amino acid Percentage Count Longest homopolymer A alanine 8.5 21 3 C cysteine 2.4 6 1 D aspartate 2.8 7 1 E glutamate 4.8 12 2 F phenylalanine 3.6 9 2 G glycine 10.1 25 1 H histidine 4.4 11 1 I isoleucine 1.2 3 1 K lysine 2.8 7 1 L leucine 19.4 48 3 M methionine 2.0 5 1 N asparagine 0.8 2 1 P proline 8.5 21 2 Q glutamine 6.5 16 1 R arginine 4.0 10 2 S serine 4.8 12 2 T threonine 4.4 11 1 V valine 4.8 12 2 W tryptophan 1.6 4 1 Y tyrosine 2.4 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342296 GPAA1 0.457 glycosylphosphatidylinositol anchor attachment protei... LOC100287119 0.072 PREDICTED: hypothetical protein XP_002342305 NFRKB 0.020 nuclear factor related to kappaB binding protein is... NFRKB 0.020 nuclear factor related to kappaB binding protein iso... PLEKHG4B 0.018 pleckstrin homology domain containing, family G (wi... PTPN23 0.016 protein tyrosine phosphatase, non-receptor type 23 [... MLL5 0.014 myeloid/lymphoid or mixed-lineage leukemia 5 MLL5 0.014 myeloid/lymphoid or mixed-lineage leukemia 5 SCXB 0.014 scleraxis homolog B SCXA 0.014 scleraxis homolog A KCNH3 0.012 potassium voltage-gated channel, subfamily H (eag-re... COL4A5 0.012 type IV collagen alpha 5 isoform 2 precursor COL4A5 0.012 type IV collagen alpha 5 isoform 1 precursor COL4A5 0.012 type IV collagen alpha 5 isoform 3 precursor SMARCD2 0.012 SWI/SNF-related matrix-associated actin-dependent r... FAM110C 0.012 hypothetical protein LOC642273 UNCX 0.012 UNC homeobox COL4A1 0.010 alpha 1 type IV collagen preproprotein XAGE5 0.010 X antigen family, member 5 NACA 0.010 nascent polypeptide-associated complex alpha subuni... GGA1 0.010 golgi associated, gamma adaptin ear containing, ARF ... SYVN1 0.010 synoviolin 1 isoform a FOXF1 0.010 forkhead box F1 GLI2 0.010 GLI-Kruppel family member GLI2 LOC100290951 0.010 PREDICTED: hypothetical protein XP_002346501 ZNF831 0.010 zinc finger protein 831 VWCE 0.010 von Willebrand factor C and EGF domains LOC338758 0.010 PREDICTED: hypothetical protein LOC338758 0.010 PREDICTED: hypothetical proteinHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
