
| Name: EXOSC6 | Sequence: fasta or formatted (272aa) | NCBI GI: 17402904 | |
|
Description: homolog of yeast mRNA transport regulator 3
|
Referenced in:
| ||
|
Composition:
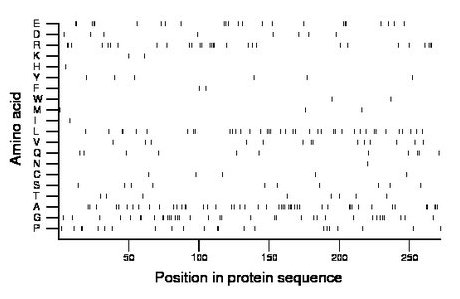
Amino acid Percentage Count Longest homopolymer A alanine 17.6 48 3 C cysteine 1.8 5 1 D aspartate 2.9 8 1 E glutamate 8.8 24 3 F phenylalanine 0.7 2 1 G glycine 13.2 36 3 H histidine 0.4 1 1 I isoleucine 0.4 1 1 K lysine 0.7 2 1 L leucine 14.3 39 3 M methionine 1.1 3 1 N asparagine 0.4 1 1 P proline 8.1 22 2 Q glutamine 4.0 11 2 R arginine 10.7 29 4 S serine 3.7 10 1 T threonine 2.9 8 1 V valine 5.1 14 2 W tryptophan 0.7 2 1 Y tyrosine 2.2 6 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 homolog of yeast mRNA transport regulator 3 LOC392145 0.271 hypothetical protein LOC392145 EXOSC4 0.160 exosome component 4 EXOSC5 0.076 exosome component Rrp46 PNPT1 0.035 polyribonucleotide nucleotidyltransferase 1 MAGI2 0.031 membrane associated guanylate kinase, WW and PDZ dom... ZCCHC3 0.025 zinc finger, CCHC domain containing 3 CHMP1A 0.023 chromatin modifying protein 1A isoform 1 LOC100288963 0.023 PREDICTED: hypothetical protein XP_002342546 LOC100292723 0.018 PREDICTED: hypothetical protein ARMC5 0.018 armadillo repeat containing 5 isoform a LOC100287558 0.018 PREDICTED: hypothetical protein XP_002343325 HCN4 0.016 hyperpolarization activated cyclic nucleotide-gated p... LOC100288796 0.016 PREDICTED: hypothetical protein LOC100288796 0.016 PREDICTED: hypothetical protein XP_002343710 ANO8 0.016 anoctamin 8 ABCB10 0.016 ATP-binding cassette, sub-family B, member 10 [Homo... LOC728767 0.016 PREDICTED: hypothetical protein LOC100290263 0.016 PREDICTED: hypothetical protein XP_002347916 LOC728767 0.016 PREDICTED: hypothetical protein RIN2 0.016 Ras and Rab interactor 2 LARP1 0.016 la related protein isoform 2 TAF10 0.014 TBP-related factor 10 LOC651986 0.014 PREDICTED: similar to forkhead box L2 LOC100288524 0.014 PREDICTED: hypothetical protein XP_002342748 LOC651986 0.014 PREDICTED: similar to forkhead box L2 MAF 0.014 v-maf musculoaponeurotic fibrosarcoma oncogene homol... MAF 0.014 v-maf musculoaponeurotic fibrosarcoma oncogene homolo... SOX1 0.014 SRY (sex determining region Y)-box 1 MYLK2 0.014 skeletal myosin light chain kinaseHuman BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
