
| Name: VAMP2 | Sequence: fasta or formatted (116aa) | NCBI GI: 172072620 | |
|
Description: vesicle-associated membrane protein 2 (synaptobrevin 2)
|
Referenced in: ER, Golgi, and the Secretory Pathway
| ||
|
Composition:
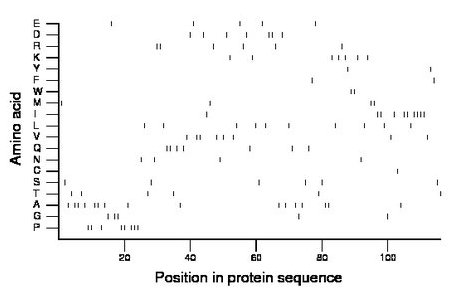
Amino acid Percentage Count Longest homopolymer A alanine 13.8 16 2 C cysteine 0.9 1 1 D aspartate 6.0 7 2 E glutamate 4.3 5 1 F phenylalanine 1.7 2 1 G glycine 4.3 5 2 H histidine 0.0 0 0 I isoleucine 8.6 10 4 K lysine 6.0 7 1 L leucine 8.6 10 1 M methionine 3.4 4 2 N asparagine 3.4 4 1 P proline 6.9 8 3 Q glutamine 6.0 7 2 R arginine 5.2 6 2 S serine 5.2 6 1 T threonine 5.2 6 1 V valine 6.9 8 2 W tryptophan 1.7 2 2 Y tyrosine 1.7 2 1 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 vesicle-associated membrane protein 2 (synaptobrevi... VAMP1 0.779 vesicle-associated membrane protein 1 isoform 1 [Homo... VAMP1 0.745 vesicle-associated membrane protein 1 isoform 2 [Hom... VAMP1 0.745 vesicle-associated membrane protein 1 isoform 3 [Homo... VAMP3 0.564 vesicle-associated membrane protein 3 VAMP4 0.279 vesicle-associated membrane protein 4 VAMP8 0.245 vesicle-associated membrane protein 8 VAMP5 0.191 vesicle-associated membrane protein 5 VAMP7 0.162 vesicle-associated membrane protein 7 isoform 2 [Ho... VAMP7 0.162 vesicle-associated membrane protein 7 isoform 1 [Homo... SEC22B 0.064 SEC22 vesicle trafficking protein homolog B UTF1 0.044 undifferentiated embryonic cell transcription factor... SGIP1 0.039 SH3-domain GRB2-like (endophilin) interacting prote... YKT6 0.034 YKT6 v-SNARE protein WASL 0.029 Wiskott-Aldrich syndrome gene-like protein LOC100129203 0.029 PREDICTED: similar to mCG50504 PRR15 0.029 proline rich 15 ZFP36L2 0.025 zinc finger protein 36, C3H type-like 2 WAS 0.025 Wiskott-Aldrich syndrome protein ZC3H10 0.025 zinc finger CCCH-type containing 10 FOXK1 0.025 forkhead box K1 FBXO41 0.025 F-box protein 41 DIAPH1 0.025 diaphanous 1 isoform 2 DIAPH1 0.025 diaphanous 1 isoform 1 LIX1L 0.025 Lix1 homolog (mouse) like LMNB1 0.020 lamin B1 LOC100293144 0.020 PREDICTED: hypothetical protein LOC100288857 0.020 PREDICTED: hypothetical protein LOC100288857 0.020 PREDICTED: hypothetical protein XP_002343308 C9orf140 0.020 tumor specificity and mitosis phase-dependent expre...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
