
| Name: LOC100131686 | Sequence: fasta or formatted (186aa) | NCBI GI: 169217622 | |
|
Description: PREDICTED: hypothetical protein isoform 2
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {186aa} PREDICTED: hypothetical protein isoform 1 | |||
|
Composition:
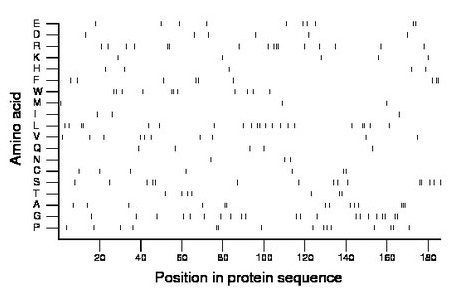
Amino acid Percentage Count Longest homopolymer A alanine 7.5 14 3 C cysteine 3.8 7 2 D aspartate 3.2 6 1 E glutamate 5.4 10 2 F phenylalanine 4.8 9 2 G glycine 11.3 21 2 H histidine 2.7 5 1 I isoleucine 1.6 3 1 K lysine 2.7 5 1 L leucine 11.3 21 2 M methionine 1.6 3 1 N asparagine 1.6 3 1 P proline 8.1 15 2 Q glutamine 2.7 5 1 R arginine 8.6 16 3 S serine 8.1 15 2 T threonine 3.8 7 2 V valine 5.4 10 1 W tryptophan 5.9 11 2 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein isoform 2 LOC100131686 1.000 PREDICTED: hypothetical protein isoform 1 LOC728701 0.060 PREDICTED: hypothetical protein MTX1 0.024 metaxin 1 isoform 1 MTX1 0.024 metaxin 1 isoform 2 LOC729575 0.016 PREDICTED: hypothetical protein LOC100288110 0.016 PREDICTED: hypothetical protein LOC100288110 0.016 PREDICTED: hypothetical protein XP_002343007 KIF26A 0.016 kinesin family member 26A C17orf96 0.013 hypothetical protein LOC100170841 GPRIN2 0.013 G protein-regulated inducer of neurite outgrowth 2 ... POU5F1B 0.013 POU class 5 homeobox 1B XKR4 0.013 XK, Kell blood group complex subunit-related family,... LOC645249 0.013 PREDICTED: hypothetical protein SDC3 0.010 syndecan 3 IRF2BP1 0.010 interferon regulatory factor 2 binding protein 1 [Ho... BBC3 0.010 BCL2 binding component 3 isoform 1 UNC84A 0.010 unc-84 homolog A isoform a MED25 0.010 mediator complex subunit 25 LOC100132345 0.008 PREDICTED: hypothetical protein HEG1 0.008 HEG homolog 1 BHMT2 0.008 betaine-homocysteine methyltransferase 2 TLE3 0.008 transducin-like enhancer protein 3 isoform a C16orf42 0.008 hypothetical protein LOC115939 TG 0.008 thyroglobulin SIGMAR1 0.008 sigma non-opioid intracellular receptor 1 isoform 1 [... DCTN1 0.008 dynactin 1 isoform 2 DCTN1 0.008 dynactin 1 isoform 1 FAM65B 0.008 hypothetical protein LOC9750 isoform 2 FAM65B 0.008 hypothetical protein LOC9750 isoform 1Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
