
| Name: FAM168B | Sequence: fasta or formatted (195aa) | NCBI GI: 112734857 | |
|
Description: hypothetical protein LOC130074
| Not currently referenced in the text | ||
|
Composition:
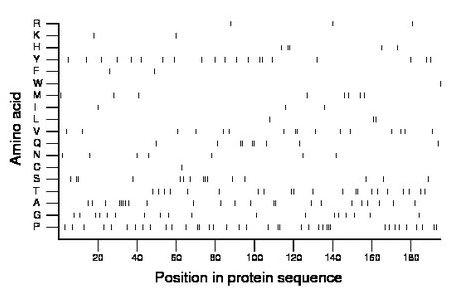
Amino acid Percentage Count Longest homopolymer A alanine 11.8 23 4 C cysteine 0.5 1 1 D aspartate 0.0 0 0 E glutamate 0.0 0 0 F phenylalanine 1.0 2 1 G glycine 9.2 18 1 H histidine 2.6 5 2 I isoleucine 1.5 3 1 K lysine 1.0 2 1 L leucine 1.5 3 2 M methionine 4.1 8 1 N asparagine 3.6 7 1 P proline 19.5 38 3 Q glutamine 4.6 9 2 R arginine 1.5 3 1 S serine 7.7 15 3 T threonine 10.8 21 2 V valine 8.2 16 2 W tryptophan 0.5 1 1 Y tyrosine 10.3 20 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 hypothetical protein LOC130074 FAM168A 0.583 hypothetical protein LOC23201 POLR2A 0.058 DNA-directed RNA polymerase II A RAPH1 0.058 Ras association and pleckstrin homology domains 1 is... MAPK1IP1L 0.056 MAPK-interacting and spindle-stabilizing protein [Ho... LGALS3 0.053 galectin 3 SMARCC2 0.053 SWI/SNF-related matrix-associated actin-dependent re... ARHGAP17 0.051 nadrin isoform 1 ARHGAP17 0.051 nadrin isoform 2 BCORL1 0.051 BCL6 co-repressor-like 1 MEGF9 0.051 multiple EGF-like-domains 9 SMARCC2 0.048 SWI/SNF-related matrix-associated actin-dependent r... LOC284297 0.048 hypothetical protein LOC284297 SMARCC2 0.045 SWI/SNF-related matrix-associated actin-dependent re... GRINA 0.043 glutamate receptor, ionotropic, N-methyl D-aspartate... GRINA 0.043 glutamate receptor, ionotropic, N-methyl D-aspartate... WBP2 0.043 WW domain binding protein 2 EWSR1 0.043 Ewing sarcoma breakpoint region 1 isoform 5 EWSR1 0.043 Ewing sarcoma breakpoint region 1 isoform 3 EWSR1 0.043 Ewing sarcoma breakpoint region 1 isoform 1 EWSR1 0.043 Ewing sarcoma breakpoint region 1 isoform 2 NACA 0.040 nascent polypeptide-associated complex alpha subuni... EIF4G1 0.038 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.038 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.038 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.038 eukaryotic translation initiation factor 4 gamma, 1 ... EIF4G1 0.038 eukaryotic translation initiation factor 4 gamma, 1 ... MED14 0.038 mediator complex subunit 14 TAF4 0.038 TBP-associated factor 4 CBLL1 0.038 Cas-Br-M (murine) ecotropic retroviral transforming...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
