
| Name: LOC646626 | Sequence: fasta or formatted (120aa) | NCBI GI: 88942880 | |
|
Description: PREDICTED: hypothetical protein
| Not currently referenced in the text | ||
Other entries for this name:
alt prot {120aa} PREDICTED: hypothetical protein alt prot {120aa} PREDICTED: hypothetical protein | |||
|
Composition:
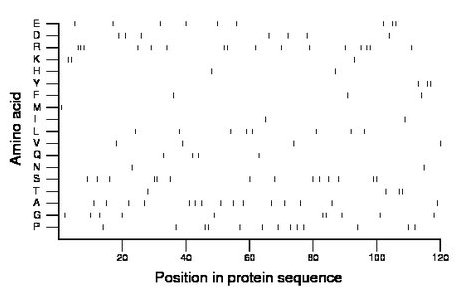
Amino acid Percentage Count Longest homopolymer A alanine 12.5 15 1 C cysteine 0.0 0 0 D aspartate 5.8 7 1 E glutamate 7.5 9 2 F phenylalanine 2.5 3 1 G glycine 8.3 10 2 H histidine 1.7 2 1 I isoleucine 1.7 2 1 K lysine 2.5 3 2 L leucine 6.7 8 1 M methionine 0.8 1 1 N asparagine 1.7 2 1 P proline 10.8 13 2 Q glutamine 3.3 4 1 R arginine 13.3 16 3 S serine 11.7 14 2 T threonine 3.3 4 2 V valine 3.3 4 1 W tryptophan 0.0 0 0 Y tyrosine 2.5 3 2 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein LOC646626 0.986 PREDICTED: hypothetical protein LOC646626 0.986 PREDICTED: hypothetical protein ZNF469 0.060 zinc finger protein 469 TCOF1 0.046 Treacher Collins-Franceschetti syndrome 1 isoform f... TCOF1 0.046 Treacher Collins-Franceschetti syndrome 1 isoform e... TCOF1 0.046 Treacher Collins-Franceschetti syndrome 1 isoform d... TCOF1 0.046 Treacher Collins-Franceschetti syndrome 1 isoform b ... TCOF1 0.046 Treacher Collins-Franceschetti syndrome 1 isoform a ... TCOF1 0.046 Treacher Collins-Franceschetti syndrome 1 isoform c ... FLJ22184 0.037 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.037 PREDICTED: hypothetical protein FLJ22184 FLJ22184 0.037 PREDICTED: hypothetical protein LOC80164 SRRM2 0.032 splicing coactivator subunit SRm300 SRRM1 0.032 serine/arginine repetitive matrix 1 CASKIN1 0.032 CASK interacting protein 1 LOC100128081 0.032 PREDICTED: hypothetical protein SART1 0.032 squamous cell carcinoma antigen recognized by T cell... CLPP 0.028 caseinolytic peptidase, ATP-dependent, proteolytic su... USP42 0.028 ubiquitin specific peptidase 42 SLC16A2 0.028 solute carrier family 16, member 2 SNIP 0.028 SNAP25-interacting protein MED1 0.023 mediator complex subunit 1 LEMD2 0.023 LEM domain containing 2 isoform 1 C10orf2 0.023 twinkle ANKRD33B 0.023 PREDICTED: ankyrin repeat domain 33B USP51 0.023 ubiquitin specific protease 51 APOB48R 0.023 apolipoprotein B48 receptor TTC15 0.018 tetratricopeptide repeat domain 15 RLTPR 0.018 RGD motif, leucine rich repeats, tropomodulin domain...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
