
| Name: LOC100288928 | Sequence: fasta or formatted (206aa) | NCBI GI: 239741926 | |
|
Description: PREDICTED: hypothetical protein XP_002342353
| Not currently referenced in the text | ||
Other entries for this name:
alt mRNA {206aa} PREDICTED: hypothetical protein | |||
|
Composition:
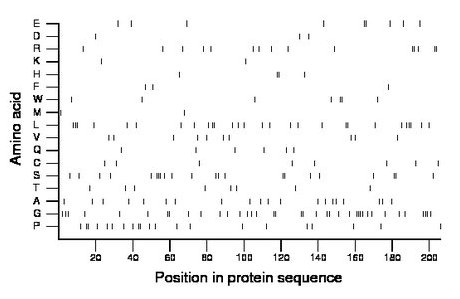
Amino acid Percentage Count Longest homopolymer A alanine 9.7 20 1 C cysteine 3.9 8 1 D aspartate 1.5 3 1 E glutamate 4.4 9 2 F phenylalanine 1.5 3 1 G glycine 18.4 38 4 H histidine 1.9 4 2 I isoleucine 0.0 0 0 K lysine 1.0 2 1 L leucine 13.6 28 3 M methionine 1.0 2 1 N asparagine 0.0 0 0 P proline 10.2 21 2 Q glutamine 2.9 6 1 R arginine 7.3 15 2 S serine 10.7 22 3 T threonine 3.9 8 1 V valine 4.9 10 1 W tryptophan 3.4 7 2 Y tyrosine 0.0 0 0 |
Comparative genomics:
Search single species RefSeq proteins at NCBI
Search summary 
Figure data | ||
Related human proteins:Protein Relative score Description Self-match 1.000 PREDICTED: hypothetical protein XP_002342353 LOC100288928 1.000 PREDICTED: hypothetical protein EP400 0.034 E1A binding protein p400 LOC100288583 0.029 PREDICTED: hypothetical protein XP_002343504 CNOT3 0.022 CCR4-NOT transcription complex, subunit 3 LOC100130600 0.019 PREDICTED: similar to hCG2014367 LOC100291533 0.019 PREDICTED: hypothetical protein XP_002347080 LOC651986 0.019 PREDICTED: similar to forkhead box L2 LOC100288002 0.019 PREDICTED: hypothetical protein XP_002342946 LOC100288524 0.019 PREDICTED: hypothetical protein XP_002342748 SNX19 0.019 sorting nexin 19 ADRA1D 0.017 alpha-1D-adrenergic receptor TRIOBP 0.017 TRIO and F-actin binding protein isoform 6 UBQLN3 0.017 ubiquilin 3 NACA 0.017 nascent polypeptide-associated complex alpha subuni... ZC3H3 0.017 zinc finger CCCH-type containing 3 YLPM1 0.017 YLP motif containing 1 LOC100288667 0.015 PREDICTED: hypothetical protein XP_002344293 KDM6B 0.015 lysine (K)-specific demethylase 6B PRR14 0.015 proline rich 14 LOC100131831 0.015 PREDICTED: hypothetical protein LOC100292287 0.015 PREDICTED: hypothetical protein LOC100131831 0.015 PREDICTED: hypothetical protein LOC100289576 0.015 PREDICTED: hypothetical protein LOC100130600 0.015 PREDICTED: similar to hCG2014367 LOC100131831 0.015 PREDICTED: hypothetical protein LOC100289576 0.015 PREDICTED: hypothetical protein XP_002343622 LOC100130600 0.015 PREDICTED: hypothetical protein LOC100130600 LOC100130600 0.015 PREDICTED: similar to hCG2014367 UTF1 0.015 undifferentiated embryonic cell transcription factor...Human BLASTP results (used to prepare the table) | |||
Gene descriptions are from NCBI RefSeq. Search results were obtained with NCBI BLAST and RefSeq entries. When identical proteins are present, the self-match may not be listed first in BLASTP output. In such cases, the table above has been reordered to place it first.
See About the Figures for the scoring system used in the figure above right. The same scoring system was used in the table of BLASTP results.
Guide to the Human Genome
Copyright © 2010 by Stewart Scherer. All rights reserved.
